Pull-Down Assays
Protein–protein interactions form the foundation of nearly all biological processes, from signal transduction and metabolic regulation to immune responses and disease progression. Pull-down assays provide a straightforward, cost-effective, and highly informative approach to identifying, verifying, and characterizing these interactions. At Creative BioMart, we offer an optimized suite of pull-down assay services—spanning GST, His-tag, MBP, and biotin-based platforms—designed to accommodate diverse research needs. Combining robust affinity purification with advanced analytical techniques such as SDS-PAGE, Western blotting, and mass spectrometry, our services deliver high-confidence interaction data to support pathway studies, target validation, and post-proteomics research.
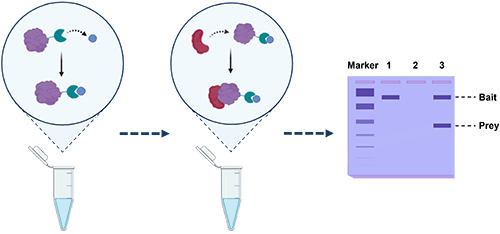
Background: Principle of Pull-Down Assays
Proteins rarely act alone. In most cellular contexts, they function as part of larger complexes or in concert with multiple interacting partners that together define their biological roles. Defining these interactions is essential for elucidating molecular mechanisms, understanding disease pathways, and developing targeted therapeutics. A variety of biochemical, biophysical, and genetic approaches exist for studying protein interactions, but many of these techniques can be complex, expensive, or limited by experimental constraints.
Pull-down assays, in contrast, offer a relatively simple and robust method for examining protein associations under conditions that closely mimic physiological interactions. The principle is straightforward: a purified “bait” protein is immobilized on an affinity matrix—typically through a fusion tag or high-specificity ligand—and then incubated with a protein mixture containing potential binding partners (“prey”). Prey proteins that exhibit affinity for the bait are captured, washed to remove nonspecific binders, and eluted for analysis.
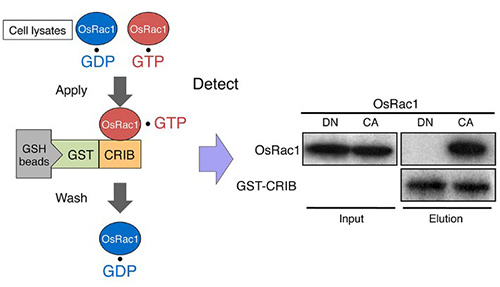
Figure 1. An example of pull-down assay: In vivo monitoring of Rac/Rop activation using a GST-CRIB pull-down assay. The CRIB domain of PAK has a high affinity for the active GTP-bound form of Rac/Rop. PAK-CRIB-binding to Rac/Rop suppresses the intrinsic and catalytic rates of GTP hydrolysis of Rac/Rop that make it possible to purify the active constitutively active (CA) form but not the inactive dominant negative (DN) form of OsRac1 from cell lysates. (Kawano et al., 2014)
The method is well suited both for discovery applications, such as identifying novel interaction partners, and validation studies, including confirmation of known interactions or mapping of binding domains. Pull-down assays can employ various tag-based affinity systems, including GST, 6xHis, MBP, and avidin–biotin arrangements. Prey proteins can originate from cell lysates, recombinant expression systems, or purified protein preparations, offering broad flexibility depending on project design.
When combined with downstream mass spectrometric analysis, pull-down assays enable high-confidence identification of interacting proteins in complex mixtures and can even support quantitative evaluation of binding strength. As an established, versatile, and economical tool in protein biochemistry, pull-down assays remain a cornerstone of protein interaction research.
Pull-Down Assays: What We Offer
Creative BioMart provides a comprehensive portfolio of pull-down assay services designed to support researchers investigating protein interactions across a wide range of biological systems. Our services include:
-
Standard Pull-Down Assays
Using tagged bait proteins immobilized on affinity beads (GST, His-tag, MBP), we capture interacting partners and analyze bound proteins via SDS-PAGE and Western blotting. This is ideal for targeted studies, validation experiments, and interaction characterization.
-
Pull-Down Assays Coupled with Mass Spectrometry
For exploratory or high-sensitivity applications, we perform LC-MS/MS–based identification of known and unknown interacting partners. This is particularly valuable in pathway mapping and identification of novel components in multi-protein complexes.
-
Biotin-Associated Pull-Down Assays
Using avidin–biotin affinity systems, our biotin-based pull-down approach enables quantitative evaluation of prey protein abundance. This method is excellent for post-proteomics studies, relative interaction quantification, or evaluation of interaction dynamics under different biological conditions.
-
Customized Assay Development
We accommodate special project needs, such as unique bait tag formats, specialized buffers, protein truncations/mutants, cofactor dependency tests, or environmental variable screening (e.g., pH, salt, temperature, inhibitors).
-
Full Analytical Support
We provide detailed data interpretation, including band identification, relative quantitation, differential analysis across experimental conditions, and recommendations for downstream validation.
Service Workflow
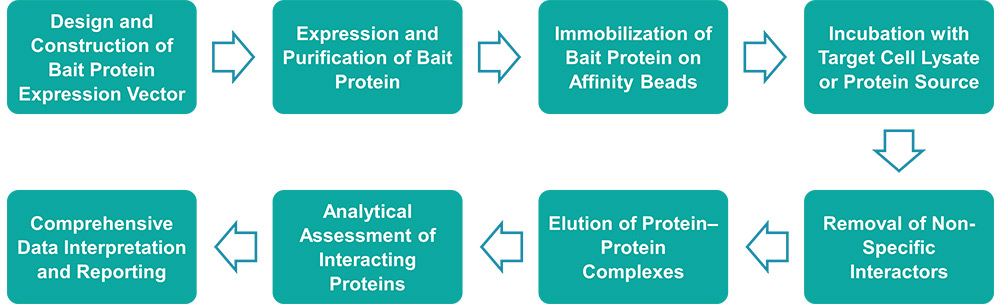
Service Features
|
Affinity Platforms Available |
Glutathione S-transferase (GST) beads |
|
Hexa-Histidine (6xHis) beads |
|
|
Maltose-binding protein (MBP) beads |
|
|
Avidin–Biotin-Based Systems |
|
|
Applications of Pull-Down Assays |
Initial screening for novel protein–protein interactions |
|
Confirmation of interactions identified by other methods |
|
|
Isolation of multi-protein complexes |
|
|
Mapping of interaction domains or key residues |
|
|
Quantitative evaluation of interaction strength |
|
|
Post-proteomics functional analysis |
What Sets Us Apart
- Highly Optimized and Reproducible Workflows: Our protocols are refined from years of experience, minimizing background binding and ensuring consistent interaction capture.
- Multiple Affinity Systems to Maximize Versatility: GST, His, MBP, and biotin-based options allow us to accommodate a wide range of bait protein formats and project requirements.
- Comprehensive Analytical Capabilities: We offer seamless integration with SDS-PAGE, Western blotting, and LC-MS/MS, enabling both targeted and discovery-driven analyses.
- Customizable Experimental Variables: Buffer composition, wash stringency, environmental conditions, and tag configurations can be tailored for challenging proteins or unique hypotheses.
- Cost-Effective Yet High-Quality Service: Pull-down assays are inherently economical, and our streamlined workflow further reduces cost while maintaining scientific rigor.
- Expert Scientific Support Throughout the Project: From experimental design to final interpretation, our team provides guidance based on extensive protein biochemistry knowledge and practical experience.
Pull-Down Assays: Case Studies
Case 1: Pull-down assays for identifying Rab protein effectors
Kawano et al., 2014. doi:10.3389/fpls.2014.00522
To uncover new effectors of the diverse mammalian Rab GTPases, researchers performed a large-scale GST pull-down screening using 60 Rab isoforms against lysates from five cell and tissue types. Mass spectrometry identified 21 Rab-binding proteins, including three GAPs—TBC1D2B, TBC1D11, and ACAP2—that interacted with specific Rabs through non-GAP domains. Notably, ACAP2 bound GTP-Rab35 via its ankyrin repeat region, influencing Rab35-dependent neurite outgrowth by modulating Arf6 activity despite lacking Rab35-GAP function. This study highlights how pull-down assays can reveal unexpected interaction mechanisms and expand understanding of Rab-mediated membrane trafficking.
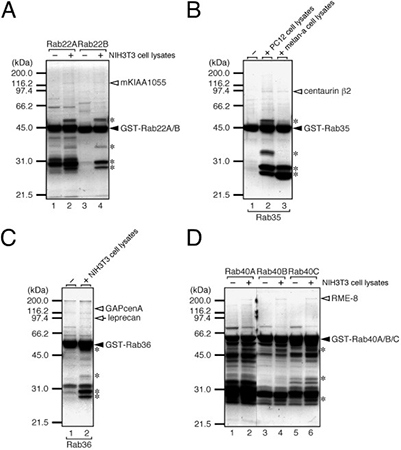
Figure 2. Isolation of specific Rab-binding proteins from cell lysates by GST pull-down assay. (A) mKIAA1055/TBC1D2B captured with GST-Rab22A/B. (B) Centaurin β2 captured with GST-Rab35. (C) GAPCenA/TBC1D11 captured with GST-Rab36. (D) RME-8 captured with GST-Rab40A/B/C. GST-Rab proteins (closed arrowheads) on glutathione beads were incubated with cell lysates or buffer controls. After washing, bound proteins were resolved by SDS-PAGE and stained. Specific candidate Rab-binding proteins (open arrowheads/arrows) were excised and identified by mass spectrometry; asterisks mark nonspecific binders. (Kawano et al., 2014)
Case 2: Pull-down assays for challenging protein–protein interactions
Bonchuk et al., 2022. doi:10.3791/64541
Traditional pull-down assays often fail when protein complexes require co-translational assembly, chaperones, or involve highly stable or unstable partners. A co-expression–coupled pull-down method overcomes these limitations by expressing differentially tagged proteins together in bacteria, eliminating separate purification steps and improving reproducibility. This approach successfully revealed interactions that conventional pull-downs could not detect, including stable ZAD dimerization and mutually exclusive binding of ENY2 with Sgf11 or CTCF fragments. It also reduced nonspecific binding compared with classic methods. Overall, co-expression pull-downs offer a faster, scalable, and more reliable strategy for studying difficult protein–protein interactions.
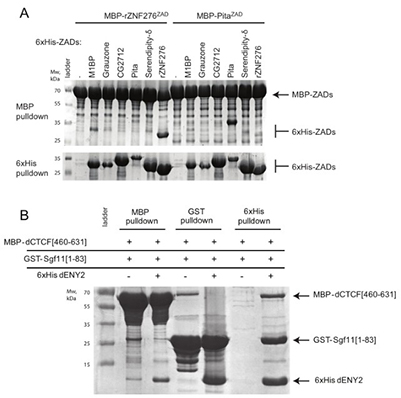
Figure 3: Representative Results. (A) Analysis of ZAD homodimerization using MBP- and 6xHis-based pull-downs. MBP- or 6xHis-tagged ZADs were co-expressed in bacteria and purified on amylose or Ni-NTA resin, then visualized by SDS-PAGE. MBP pull-downs show dimerization; 6xHis pull-downs are shown only as expression controls. (B) Assessment of mutually exclusive binding of ENY2 with Sgf11 or CTCF. GST-Sgf11, MBP-CTCF, and 6xHis-ENY2 were co-expressed and purified with the corresponding resins, followed by SDS-PAGE detection. (Bonchuk et al., 2022)
Pull-Down Assays: Customer Feedback
“We engaged Creative BioMart to identify novel protein interactors for a kinase involved in inflammatory signaling. Their team expertly prepared GST-tagged bait constructs, optimized the pull-down conditions, and coupled the assay with mass spectrometry. The resulting dataset revealed several previously unknown binding partners, which are now the focus of our follow-up functional studies. The technical insight and speed of delivery were outstanding.”
— R&D Director, Inflammation Biology | Global Pharmaceutical Company
“Our lab needed to confirm domain-specific interactions between a transcription factor and its co-regulator. Creative BioMart performed MBP pull-down assays using truncation constructs and provided clear, quantitative Western blot data. The precision of their assays allowed us to map the critical interaction domain accurately, accelerating our publication timeline and downstream mechanistic experiments.”
— Principal Investigator | Academic Research Institute
“We required quantitative analysis of protein complexes affected by oxidative stress in endothelial cells. Creative BioMart’s biotin-associated pull-down assay not only captured the complex efficiently but also allowed us to measure relative changes in interacting protein abundance. Their detailed analysis and recommendations significantly advanced our understanding of stress-dependent signaling pathways.”
— Senior Scientist, Cardiovascular Research Unit | Biotech Company
“For our structural biology project, we needed to isolate a multi-protein complex in sufficient purity for cryo-EM. Creative BioMart’s His-tag pull-down workflow successfully captured the full complex from mammalian cell lysates while preserving integrity. Their team provided mass spectrometry confirmation and troubleshooting support, which was crucial for moving the project to structural analysis.”
— Head of Structural Biology | Pharmaceutical Research Lab
Pull-Down Assays Service: Frequently Asked Questions (FAQs)
-
Q: What types of proteins can be analyzed with pull-down assays?
A: Pull-down assays are versatile and can accommodate a wide range of proteins, including full-length, truncated, or tagged constructs. Bait proteins can be expressed in bacterial, mammalian, insect, or cell-free systems, while prey proteins can come from cell lysates, recombinant expression, or purified protein preparations. This flexibility makes the assay suitable for both targeted studies and discovery-driven projects. -
Q: Which affinity tags and bead systems do you offer?
A: We provide multiple tag options to suit different experimental needs: GST, His-tag (6xHis), MBP, and biotin-based systems. These tags are compatible with high-quality affinity beads, ensuring stable immobilization and efficient capture of interacting partners. Biotin-associated pull-down assays additionally allow for quantitative evaluation of binding. -
Q: How do you ensure specificity and reproducibility?
A: We use optimized washing and incubation conditions to minimize nonspecific binding while preserving physiologically relevant interactions. Bait proteins are carefully expressed and purified to maintain native conformation, and our workflows are designed for reproducibility across multiple experiments and sample types. -
Q: Can pull-down assays detect novel protein–protein interactions?
A: Yes. Pull-down assays can serve both as a discovery tool and a validation platform. When coupled with mass spectrometry, they enable identification of previously unknown interactors, providing insights into signaling pathways, multi-protein complexes, and functional networks. -
Q: How are interactions analyzed after the pull-down?
A: Eluted complexes can be analyzed using SDS-PAGE, Western blotting, or high-resolution mass spectrometry, depending on project goals. For quantitative studies, biotin-associated assays allow assessment of relative binding levels. A detailed report with data interpretation and recommendations for follow-up experiments is provided. -
Q: How do pull-down assays compare with other protein interaction methods?
A: Pull-down assays are simple, cost-effective, and highly flexible. Unlike some genetic or high-throughput platforms, they do not require live cell screening and can handle proteins that are difficult to express in other systems. When combined with mass spectrometry, they offer both discovery and validation capabilities at a lower cost than many alternative methods. -
Q: Can the workflow be customized for special project needs?
A: Absolutely. Our team can adjust tag type, buffer composition, wash stringency, elution conditions, and protein source to meet project-specific requirements, including challenging proteins, low-abundance targets, or multi-protein complex studies. -
Q: What support is provided for data interpretation?
A: Beyond generating raw data, we provide expert analysis, including identification of interaction partners, relative quantitation, and guidance on functional relevance. We also advise on validation strategies and potential follow-up experiments to strengthen your study.
Other Resources
Related Services
- Protein Interaction Service
- Membrane-Based Yeast Two-Hybrid System
- Mammalian Two-Hybrid System
- Co-Immunoprecipitation (Co-IP) Service
- Phage Display Platform
Related Products
References:
- Bonchuk A, Zolotarev N, Balagurov K, Arkova O, Georgiev P. Pulldown assay coupled with co-expression in bacteria cells as a time-efficient tool for testing challenging protein-protein interactions. JoVE. 2022;(190):64541. doi:10.3791/64541
- Kawano Y, Kaneko-Kawano T, Shimamoto K. Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci. 2014;5. doi:10.3389/fpls.2014.00522
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

