Protein Localization: Compare the Microscopy-based and Fluorescent Protein Tagging Method
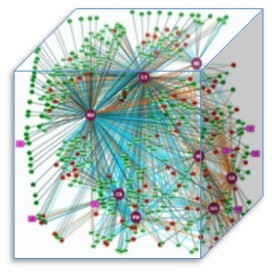
The picture shows the experimentally-determined subcellular localization of almost 500 human proteins using both immunofluorescence and GFP-tagging.
Microscopy-based method and fluorescent protein tagging method are two of the most widely used approaches to determine protein localization. But the question that which one is better has argued for decades.
Where each protein locates in cell is a key factor for us to understand how cell works.
In order to figure the issue, recently scientists did an experimental study to compare the two methods. In their study, each protein was assigned to a localisation class (CY, cytoplasm; CSK, cytoskeleton; ER, endoplasmic reticulum; GO, Golgi apparatus; MI, mitochondria; NU, nucleus; PM, plasma membrane; VE, vesicle). As the picture shows, each node represents a protein, with dark green nodes indicating identical localizations recorded between the two methods, light green nodes indicating similar localizations between the two methods, and red nodes indicating conflicting localizations between the two methods. They wished to distinguish the standard methodology for ultimately determining the localization of the entire human proteome. They checked 500 proteins, and 50% of the proteins haven’t been clarified in their localization annotation.
After the experiment, they found both antibody-based and fluorescent-tagging methods are highly complementary to each other, and show a high degree of correlation for the determination of protein localization in cells if they adhere to specific rules. The findings also provide a significant and experimentally validated data set of protein localization in mammalian cells.
The study solved a long argued issue and offers a valuable resource for the scientific community.
Article Link: Protein Localization: Compare the Microscopy-based and Fluorescent Protein Tagging Method

