Heme Metabolism
Related Symbol Search List
Immunology Background
About Heme Metabolism
Heme metabolism refers to the processes involved in the synthesis, degradation, and recycling of heme, a vital molecule found in various tissues and cells of the body. Heme is a complex molecule composed of a porphyrin ring structure with an iron ion at its center. It plays a crucial role in several biological processes, including oxygen transport, energy production, and enzymatic reactions. Here's an introduction to heme metabolism and its related molecules:
Heme Synthesis: Heme synthesis, also known as heme biosynthesis, occurs primarily in the liver and bone marrow. It involves a series of enzymatic reactions known as the heme biosynthetic pathway. The pathway starts in the mitochondria and continues in the cytoplasm. Key steps include the synthesis of aminolevulinic acid (ALA) from glycine and succinyl-CoA, and the subsequent conversion of ALA into porphobilinogen (PBG). Several enzymatic steps and intermediates follow, ultimately leading to the formation of heme.
Heme Degradation: Heme degradation, also known as heme catabolism, occurs mainly in the reticuloendothelial system, primarily in the liver and spleen. The process involves the breakdown of heme into biliverdin, carbon monoxide (CO), and iron. The enzyme heme oxygenase catalyzes the conversion of heme into biliverdin, which is then further converted to bilirubin. Bilirubin is transported to the liver, conjugated with glucuronic acid, and excreted in bile as bilirubin diglucuronide.
Heme Transport and Binding Proteins: Heme requires transport proteins to facilitate its movement within the body. Heme carrier proteins, such as HRG-1 (heme-responsive gene 1) and FLVCR (feline leukemia virus subgroup C receptor), are involved in heme uptake and transport across cell membranes. Inside cells, heme can bind to various proteins, including hemoproteins such as hemoglobin and myoglobin, cytochromes, and enzymes involved in heme metabolism.
Heme Oxygenases: Heme oxygenases are enzymes involved in heme degradation. They catalyze the conversion of heme into biliverdin, releasing iron and carbon monoxide (CO) as byproducts. There are two isoforms of heme oxygenase: heme oxygenase-1 (HO-1) and heme oxygenase-2 (HO-2). HO-1 is an inducible form that responds to various stimuli, such as oxidative stress and inflammation, while HO-2 is constitutively expressed.
Iron Metabolism: Heme metabolism is closely linked to iron metabolism since heme contains an iron ion at its core. Iron is essential for heme synthesis, and heme degradation releases iron, which can be recycled for further use. Iron regulatory proteins (IRPs) and iron-responsive elements (IREs) play a crucial role in regulating iron homeostasis and its availability for heme synthesis.
Heme-Containing Proteins: Heme is a critical component of various proteins, known as hemoproteins, that perform essential biological functions. Examples of heme-containing proteins include hemoglobin (oxygen transport in red blood cells), myoglobin (oxygen storage in muscle cells), cytochromes (electron transport in mitochondria), and catalases (involved in antioxidant defense).
Understanding heme metabolism and its related molecules is crucial for comprehending the regulation of heme synthesis, degradation, and its role in various biological processes. Dysregulation of heme metabolism can lead to disorders such as porphyrias (defects in heme synthesis), hemolytic anemias (increased heme degradation), and iron overload conditions. Additionally, heme metabolism and heme-containing proteins are potential targets for therapeutic interventions and drug development.
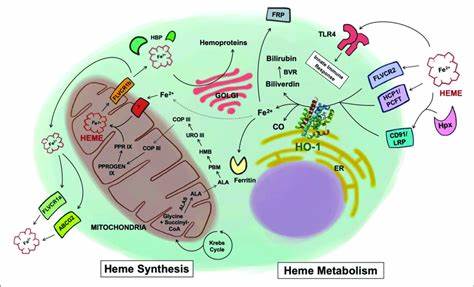
Physiological Functions of Heme Metabolism
The physiological functions of heme metabolism and related molecules are diverse and essential for the proper functioning of the body. Here are some key physiological functions:
Oxygen Transport: One of the primary functions of heme is to bind and transport oxygen. Hemoglobin, a heme-containing protein found in red blood cells, binds to oxygen in the lungs and carries it to tissues throughout the body. This enables the oxygenation of tissues and organs, supporting their metabolic activities.
Oxygen Storage: Myoglobin, another heme-containing protein, is responsible for oxygen storage in muscle cells. It binds oxygen in areas of high oxygen concentration (e.g., during breathing) and releases it when oxygen levels are low, ensuring a steady supply of oxygen for muscle activity.
Electron Transport: Heme is a crucial component of cytochromes, which are involved in electron transport chains in mitochondria. Cytochromes accept and donate electrons during cellular respiration, facilitating the production of adenosine triphosphate (ATP), the energy currency of cells.
Enzymatic Reactions: Heme-containing enzymes, known as heme enzymes or hemoproteins, play a vital role in various enzymatic reactions. For example, cytochrome P450 enzymes are involved in the metabolism of drugs, toxins, and endogenous compounds. Other heme enzymes, such as catalase and peroxidase, are involved in antioxidant defense and the breakdown of reactive oxygen species.
Bilirubin Metabolism: Heme metabolism is intricately linked to the production and metabolism of bilirubin. Bilirubin is produced during heme degradation and is further metabolized in the liver. It serves as an important marker for liver function and is excreted in bile. Proper bilirubin metabolism is crucial for the removal of waste products from the body.
Iron Homeostasis: Heme metabolism is connected to iron metabolism. Heme synthesis requires iron, and heme degradation releases iron. Iron is essential for various physiological processes, including oxygen transport, DNA synthesis, and cellular respiration. Heme metabolism and iron homeostasis are tightly regulated to ensure an adequate supply of iron for heme synthesis and other cellular functions.
Antioxidant Defense: Heme oxygenase, an enzyme involved in heme degradation, produces biliverdin and carbon monoxide as byproducts. Biliverdin is further converted to bilirubin, which has antioxidant properties. This antioxidant effect helps protect cells from oxidative stress and reduces tissue damage caused by reactive oxygen species.
Cellular Signaling: Heme can act as a signaling molecule in various cellular processes. For example, heme can regulate gene expression through interactions with specific transcription factors, such as the heme-regulated inhibitor (HRI) and the nuclear receptor Rev-Erb. Heme can also modulate cellular processes by affecting the activity of certain signaling pathways.
Understanding the physiological functions of heme metabolism and related molecules is crucial for comprehending normal cellular and tissue function. Dysregulation or defects in heme metabolism can lead to various disorders, including anemias, porphyrias, and oxidative stress-related conditions. Studying heme metabolism provides insights into the mechanisms underlying these disorders and offers opportunities for therapeutic interventions aimed at restoring normal heme metabolism and function.
Available Resources for Heme Metabolism
Creative BioMart delivers a diverse range of products and customized services to aid in heme metabolism research. Our offerings encompass recombinant proteins, cell and tissue lysates, protein pre-coupled beads, antibodies, and more, all aimed at unraveling the mechanisms governing their functions.
Moreover, we provide comprehensive resource support, spanning pathways, protein functionalities, interacting proteins, associated research areas, relevant articles, and other materials about heme metabolism. This expansive support is tailored to enhance understanding and delve into the functions and regulatory mechanisms of these pivotal molecules.
Explore the molecules related to heme metabolism below for a more comprehensive resource.
We are dedicated to providing you with high-quality research tools and services to help you achieve successful scientific outcomes. If you have any further questions or require custom services, please feel free to contact us at any time.
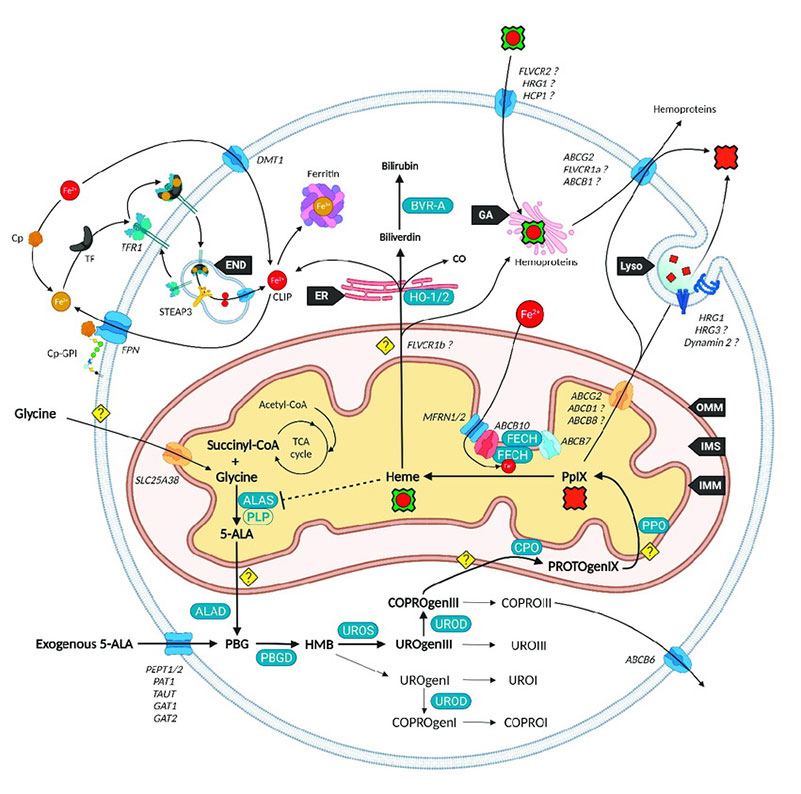 Fig.2 Current vision of the major players of heme metabolism and their interactions. (Kiening M, et al., 2022)
Fig.2 Current vision of the major players of heme metabolism and their interactions. (Kiening M, et al., 2022)
References:
- Lee GR, Shaefi S, Otterbein LE. HO-1 and CD39: It Takes Two to Protect the Realm. Front Immunol. 2019;10:1765. Published 2019 Jul 26.
- Kiening M, Lange N. A Recap of Heme Metabolism Towards Understanding Protoporphyrin IX Selectivity in Cancer Cells. Int J Mol Sci. 2022;23(14):7974. Published 2022 Jul 19.

