ISWI Family
Related Symbol Search List
Immunology Background
Background
The ISWI (Imitation Switch) family is one of the best conserved ATPase families. It possesses a highly conserved SWI2/SNF2 family ATPase domain, belonging to the superfamily of DEAD/H-helicases, that provides the motor for chromatin remodeling.
Structure of ISWI Family
ISWI family proteins have a central ATPase structural domain, which provides energy for chromatin remodeling through the hydrolysis of ATP. ATPase subunits have a characteristic HAND-SANT-SLIDE domains with DNA binding activity, which are involved in the recognition of specific histone modifications and DNA sequences, and which contribute to the interaction with histones and DNA to achieve nucleosome sliding. ISWI complexes are often composed of additional subunits that help regulate their activity and targeting specificity. These accessory subunits may vary depending on the particular complex and its function. ISWI complexes contain regions that interact with nucleosomes (components of chromatin). These interactions allow the complex to slide, space, or remodel nucleosomes along the DNA.
The ISWI Complex: Types and Composition
In mammals, a specific ISWI complex is composed of one ATPase subunit (SMARCA5 or SMARCA1) and one to three noncatalytic subunits. Specially, both human ATPases, SMARCA1 and SMARCA5, form stable complexes with all regulatory subunits, which expands the ISWI complex members up to 16, including RSF-1/RSF-5 (SMARCA1/5 and RSF1), ACF-1/ACF-5 (SMARCA1/5 and BAZ1A), CHRAC-1/CHRAC-5 (SMARCA5/1, BAZ1A, CHRAC1 and POLE3), WICH-1/WICH-5 (SMARCA1/5 and BAZ1B), NoRC-1/NoRC-5 (SMARCA1/5 and BAZ2A), NuRF-1/NuRF-5 (SMARCA1/5, BPTF, RBBP7 and RBBP4), CERF-1/CERF-5 (SMARCA1/5 and CECR2) and BRF-1/BRF-5 (SMARCA1/5, BAZ2B). Different subunits possess different functional domains and play distinct roles in complexes.
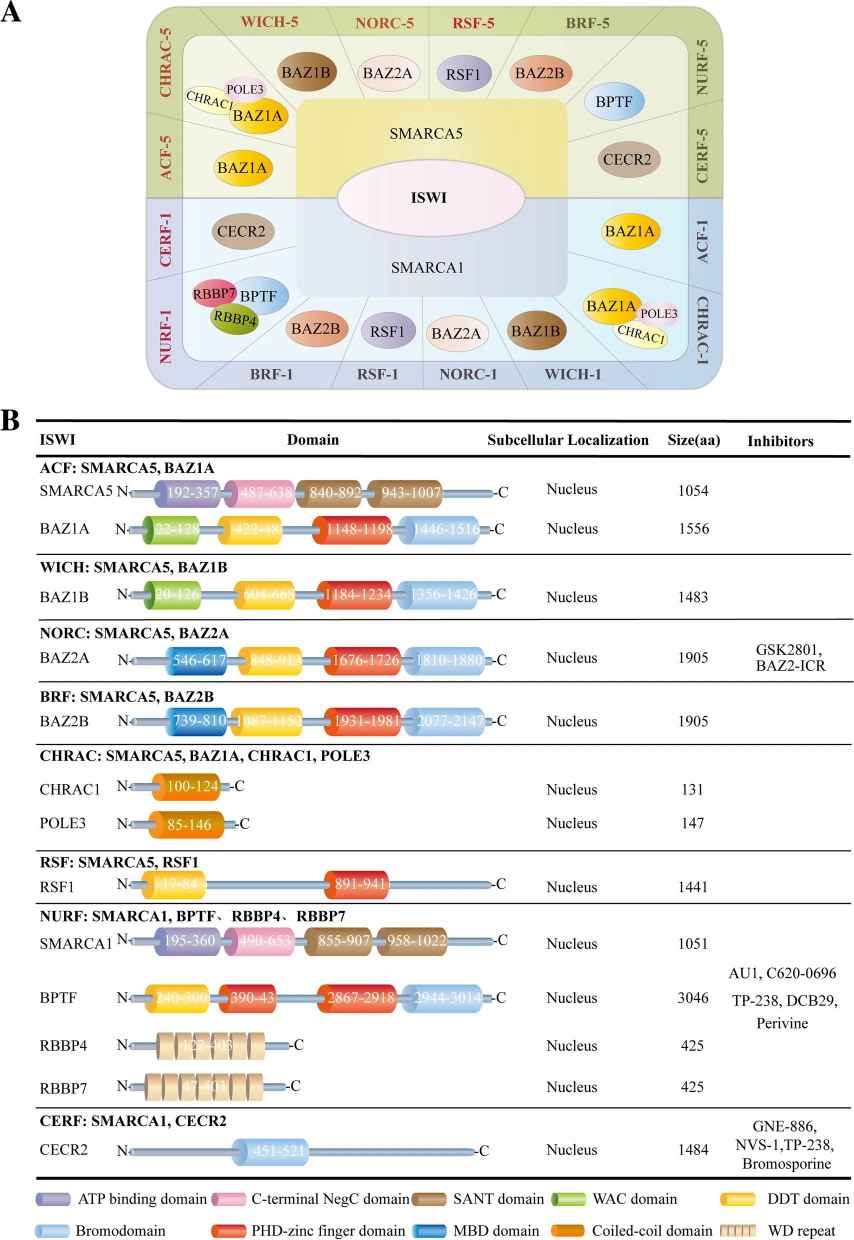 Fig.1 Classification of ISWI family. Molecular components, functional domains, subcellular localization, and targeting inhibitors for the ISWI family. (Li, Y., et al., 2021)
Fig.1 Classification of ISWI family. Molecular components, functional domains, subcellular localization, and targeting inhibitors for the ISWI family. (Li, Y., et al., 2021)Functions of ISWI Family
The ISWI family of chromatin remodelers plays a significant role in various biological processes, including cell differentiation, regulation of gene expression, and DNA repair. Here's how ISWI family members are involved in each of these processes:
| Functions | Details |
|---|---|
| Chromatin Remodeling |
Role: ISWI family members are key players in chromatin remodeling, which involves altering the structure of chromatin to regulate gene expression. Mechanism: They use ATP hydrolysis to move, reposition, and reorganize nucleosomes along DNA, influencing the accessibility of DNA to transcription factors and other regulatory proteins. Importance: This process is vital for gene regulation, DNA repair, replication, and maintenance of chromatin structure. |
| Cell Differentiation |
Role: ISWI family chromatin remodelers are crucial for cell differentiation, the process by which unspecialized cells acquire specific functions and characteristics. Mechanism: During cell differentiation, ISWI remodelers help regulate gene expression patterns by modifying chromatin structure. They can create regions of open chromatin that allow for the activation of cell type-specific genes while silencing genes that are not required for the differentiated cell's function. Outcome: Proper chromatin remodeling by ISWI complexes is essential for ensuring that cells differentiate into the correct cell types and maintain their specialized functions. |
| Regulation of Gene Expression |
Role: ISWI family members are key players in regulating gene expression by modulating chromatin structure to control access to DNA for transcription factors and other regulatory proteins. Mechanism: ISWI remodelers use ATP hydrolysis to slide nucleosomes along DNA, altering the spacing and positioning of nucleosomes. This activity can either promote gene activation by making the DNA more accessible or repress gene expression by compacting chromatin. Outcome: By dynamically regulating chromatin structure, ISWI remodelers fine-tune gene expression patterns in response to cellular signals and environmental cues, ensuring the proper functioning of the cell. |
| DNA Replication and Repair |
Role: ISWI family members are involved in maintaining chromatin structure during DNA replication and facilitating DNA repair processes. Mechanism: They regulate chromatin organization to ensure accurate DNA replication and repair by facilitating access to damaged DNA sites. Importance: Proper chromatin remodeling is essential for maintaining genomic stability and preventing mutations that can lead to diseases like cancer. |
| Developmental Processes |
Role: ISWI remodelers are critical for normal development and differentiation during embryogenesis and tissue formation. Mechanism: They regulate the expression of genes involved in developmental processes by modulating chromatin structure. Significance: The proper function of the ISWI family is necessary for orchestrating the complex gene regulatory networks that drive development. |
| Epigenetic Regulation |
Role: ISWI family members contribute to epigenetic regulation by establishing and maintaining specific chromatin states associated with gene expression patterns. Mechanism: They help propagate and inherit epigenetic marks across cell divisions, influencing gene expression and cellular function. Importance: Epigenetic regulation by ISWI remodelers is crucial for cellular memory, differentiation, and response to environmental cues. |
Members and Features of ISWI Family
The ISWI family includes several members that play crucial roles in regulating chromatin structure, gene expression, and various cellular processes. Here are some of the key members of the ISWI family along with their features and functions:
| Members | Features and function |
|---|---|
| SNF2H (SMARCA5) |
Features: Contains an ATPase domain and additional structural motifs like chromodomains. Function: Involved in nucleosome sliding and spacing, regulating gene expression, DNA repair, and chromatin organization during processes like DNA replication and transcription. |
| SNF2PH (SMARCA5) |
Features: Similar to SNF2H with ATPase and chromatin-binding domains. Function: Participates in chromatin remodeling, gene regulation, and maintenance of chromatin structure critical for various cellular processes. |
| WSTF (SMARCA5) |
Features: Contains an ATPase domain and specific chromatin-binding domains. Function: Plays a role in chromatin remodeling, regulation of gene expression, and maintenance of chromatin structure essential for proper cellular function. |
| SNF2L (SMARCA1) |
Features: Shares similarities with SNF2H, with an ATPase domain and chromodomains. Function: Plays a role in chromatin remodeling, gene regulation, and maintenance of chromatin structure critical for gene expression and cell function. |
| ACF/CHRAC |
Features: Complexes containing ISWI subunits with ATPase activity. Function: Involved in nucleosome assembly and positioning, chromatin organization, gene regulation, and maintenance of genomic stability during DNA replication and repair processes. |
| RSF (Remodeling and Spacing Factor) |
Features: Contains an ATPase subunit and additional protein domains for chromatin binding. Function: Involved in nucleosome spacing, regulation of gene expression, and chromatin organization during processes like DNA replication and repair. |
| NoRC (Nucleolar Remodeling Complex) |
Features: Contains ISWI subunits and additional components for nucleolar organization. Function: Involved in nucleolar structure maintenance, gene silencing, and modulation of ribosomal RNA expression through chromatin remodeling. |
| EP400 |
Features: ATPase domain and various chromatin-binding modules. Function: Plays a role in chromatin remodeling, gene regulation, and maintenance of chromatin structure essential for diverse cellular processes such as DNA repair and transcriptional regulation. |
These are just some examples of the members of the ISWI family that, together, form the ISWI family and are involved in regulating chromatin structure and function, as well as regulating gene expression and cellular processes. Each member plays a specific role in the complex, working together in harmony to maintain genome stability and proper function within the cell.
ISWI Family in Disease Onset and Progression
Here is an exploration of the differential expression and regulatory mechanisms of ISWI family members in disease onset and progression:
Cancer
- Alterations in ISWI family member expression levels have been linked to various cancers. For instance, overexpression of SNF2H (SMARCA5) has been observed in breast cancer, while reduced expression is associated with lung cancer.
- In breast cancer tumors, the overexpression of SNF2H (SMARCA5) contributes to tumor growth and metastasis by altering gene expression profiles associated with cell proliferation and invasion. This overexpression can also lead to aberrant chromatin remodeling, activating oncogenes and silencing tumor suppressor genes, which further drive the progression of breast cancer.
Neurological Disorders
- Studies have shown differential expression of ISWI family members in neurological disorders like Alzheimer's disease and Parkinson's disease. Changes in the expression of these remodelers can impact neuronal gene expression and function.
- For example, in Alzheimer's disease brains, members of the ACF/CHRAC complex show differential expression levels, affecting the regulation of genes related to neuronal function and synaptic plasticity. This dysregulation of chromatin remodeling by the complex can impact the expression of genes linked to neurodegeneration and cognitive decline in Alzheimer's disease.
Developmental Disorders
- Dysregulation of ISWI family members has been implicated in developmental disorders such as Cornelia de Lange syndrome and Coffin-Siris syndrome.
- Cornelia de Lange Syndrome is characterized by reduced expression of WSTF (SMARCA5), which plays a key role in regulating genes crucial for development and growth. This decrease in WSTF expression can cause chromatin structure abnormalities, impacting the expression of genes necessary for proper development and contributing to the syndrome's phenotype.
Cardiovascular Diseases
- Changes in the expression of ISWI family members have been reported in cardiovascular diseases like heart failure and atherosclerosis. ISWI remodelers influence the expression of genes involved in cardiovascular function and disease progression, impacting processes like cardiac remodeling, inflammation, and vascular health.
- The altered expression of SNF2H in atherosclerotic plaques plays a role in regulating genes related to vascular inflammation and plaque stability. Through chromatin remodeling, SNF2H can affect the expression of genes involved in endothelial dysfunction and inflammatory responses, contributing to the development of atherosclerosis.
Immune Disorders
- ISWI family members are involved in immune cell differentiation and function. Dysregulation of ISWI-mediated chromatin remodeling can impact immune cell gene expression profiles, leading to immune dysfunction and inflammatory responses seen in various disorders.
- In patients with Systemic Lupus Erythematosus (SLE), dysregulated expression of ISWI family members in immune cells affects transcriptional regulation of immune-related genes. This altered chromatin remodeling mediated by ISWI disrupts immune cell function and cytokine production, potentially contributing to the development of SLE and autoimmune responses.
In summary, the differential expression and regulatory mechanisms of ISWI family members in disease onset and progression highlight their importance in various pathological conditions. Dysregulation of these chromatin remodelers can disrupt gene expression patterns, chromatin structure, and cellular processes, contributing to the development and progression of diseases across different biological systems. Targeting ISWI family members could offer new avenues for therapeutic interventions in diseases where their dysregulation plays a significant role.
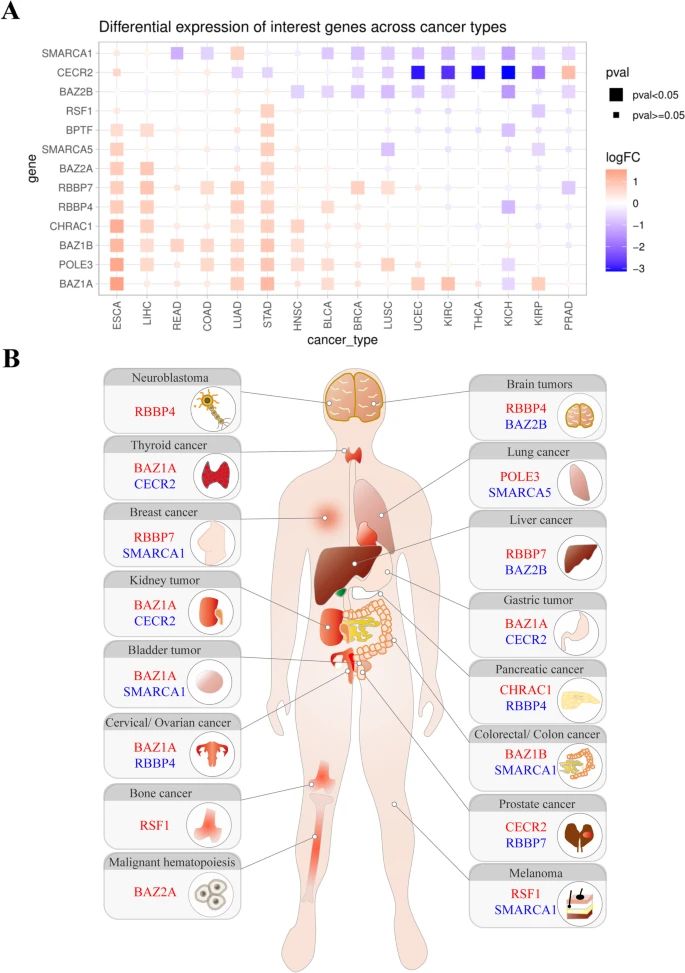 Fig.2 Gene expression analysis of ISWI family members in human cancers. (Li, Y., et al., 2021)
Fig.2 Gene expression analysis of ISWI family members in human cancers. (Li, Y., et al., 2021)Case Study
Case 1: Gong NN, Dilley LC, Williams CE, et al. The chromatin remodeler ISWI acts during Drosophila development to regulate adult sleep. Sci Adv. 2021;7(8):eabe2597.
Sleep disruptions are commonly reported symptoms in neurodevelopmental disorders, but the mechanisms linking brain development to normal sleep are not well understood. A study using Drosophila found that the chromatin remodeler Imitation SWItch/SNF (ISWI) is necessary for adult fly sleep. Loss of ISWI also affects circadian rhythms, memory, and social behavior in flies, with ISWI playing different roles in different cells and during different developmental stages. ISWI expression in type I neuroblasts is essential for adult sleep and the formation of a brain region associated with learning. Flies expressing human ISWI homologs SMARCA1 and SMARCA5 show varying degrees of rescue of adult behaviors, while SMARCA5 variants in patients with NDDs do not improve sleep. The study suggests that sleep disturbances in NDDs may have their roots in early development, and highlights the importance of chromatin remodeling machinery in the formation of sleep circuits.
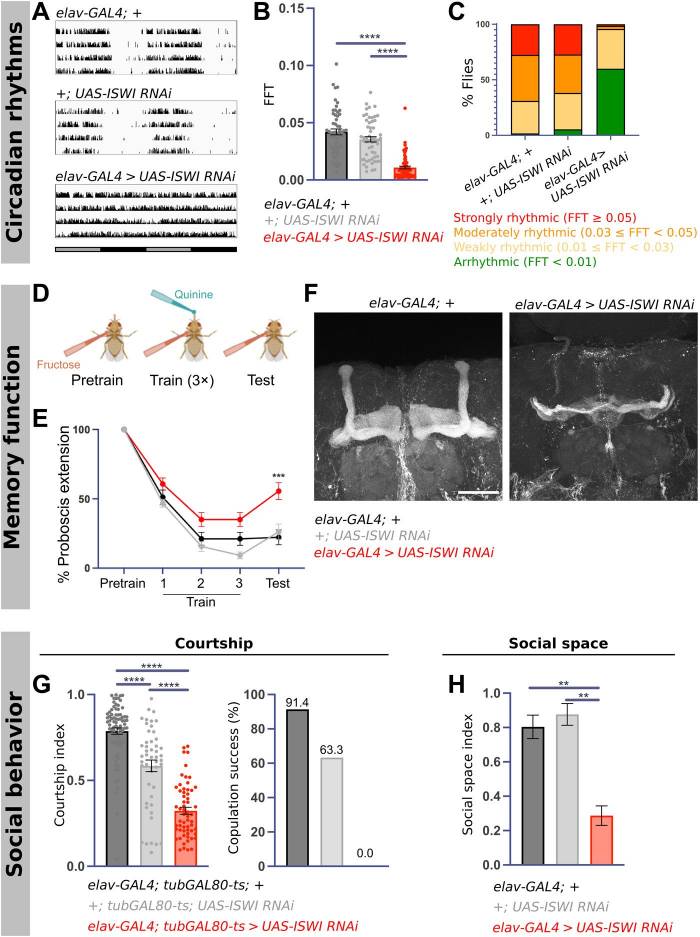 Fig.1 ISWI knockdown results in circadian arrhythmicity, memory deficits, and social dysfunction.
Fig.1 ISWI knockdown results in circadian arrhythmicity, memory deficits, and social dysfunction.Case 2: Li D, Wang Q, Gong NN, et al. Pathogenic variants in SMARCA5, a chromatin remodeler, cause a range of syndromic neurodevelopmental features. Sci Adv. 2021;7(20):eabf2066.
Intellectual disability (ID) is a complex neurodevelopmental disorder that can be caused by various genetic factors. Despite extensive research, the molecular mechanisms underlying over 50% of cases remain unknown. A recent study identified mutations in the SMARCA5 gene, which encodes the ATPase motor of the ISWI chromatin remodeler, as a novel cause of neurodevelopmental disorder. Twelve individuals with rare heterozygous variants in SMARCA5 were found to exhibit mild developmental delay, postnatal short stature, microcephaly, and distinct facial features. Further experiments in fruit flies revealed that loss of Iswi function led to developmental abnormalities in body size, dendrite complexity, and neural function. Importantly, these defects were rescued by wild-type SMARCA5, highlighting the role of this gene in neurodevelopment. Overall, these findings illustrate how mutations in SMARCA5 can contribute to a neurodevelopmental syndrome characterized by mild facial dysmorphia.
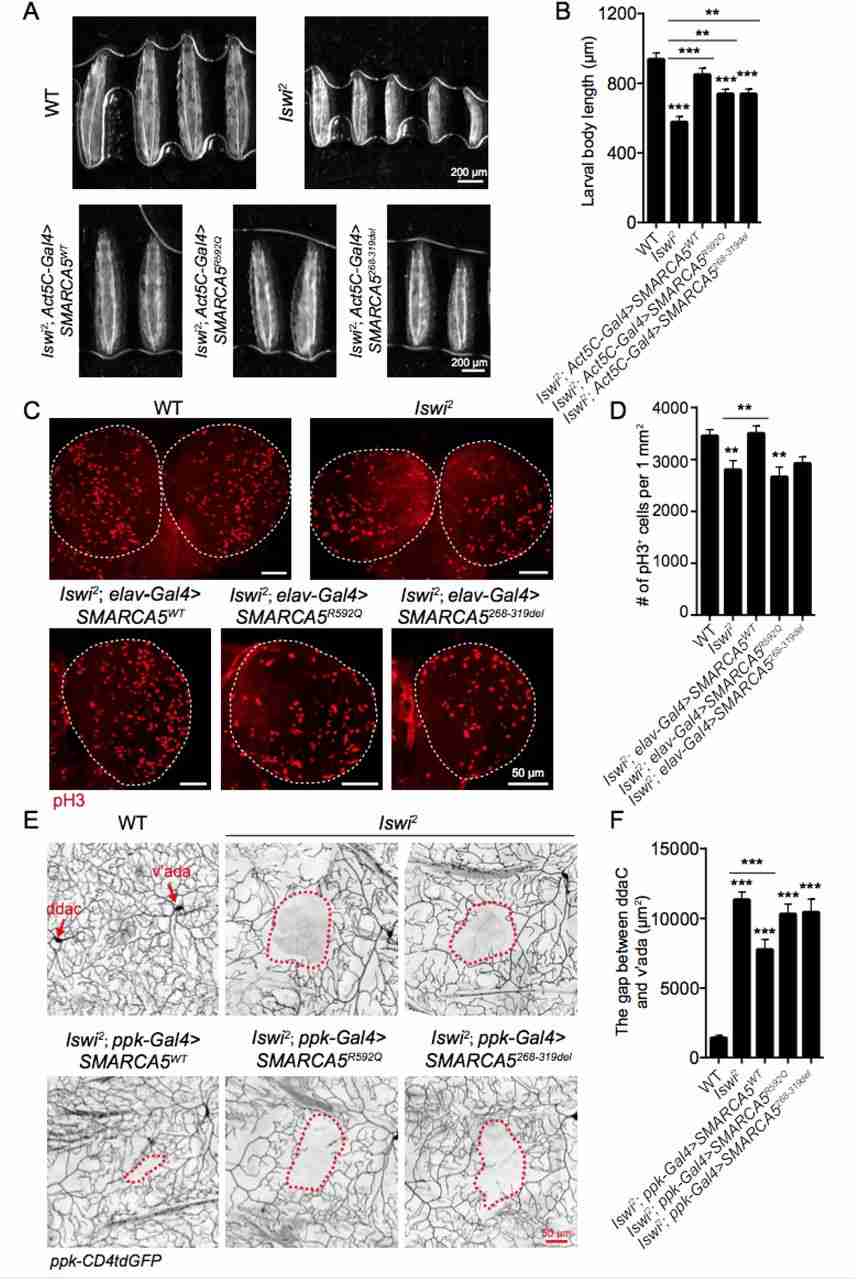 Fig.2 Iswi loss of function leads to developmental defects in Drosophila larvae.
Fig.2 Iswi loss of function leads to developmental defects in Drosophila larvae.Related References
- Li, Y., Gong, H., Wang, P. et al. The emerging role of ISWI chromatin remodeling complexes in cancer. J Exp Clin Cancer Res 40, 346 (2021).
- Oppikofer M, Bai T, Gan Y, et al. Expansion of the ISWI chromatin remodeler family with new active complexes. EMBO Rep. 2017;18(10):1697-1706.
- Bartholomew B. ISWI chromatin remodeling: one primary actor or a coordinated effort. Curr Opin Struct Biol. 2014;24:150-155.

