Protocol of Affinity Enrichment of Phosphorylated Proteins
The important role of phosphorylated protein in cell life activities has made it a hotspot in proteomics research. However, the content of phosphorylated proteins in organisms is very low, and only a few proteins phosphorylate under specific stimulation; Secondly, phosphorylation modification has variability, which makes the protein under different conditions have different phosphorylation forms, and the existing analysis methods lack dynamic analysis of phosphorylation sites, making it more difficult to identify phosphorylation sites. Therefore, the enrichment of phosphorylated proteins and the improvement of the relative content of phosphorylated proteins have become the important content of phosphorylated proteomics research.
At present, many commercial kits are made based on the principle of affinity enrichment of phosphorylated proteins. The enrichment of phosphorylated proteins is realized by the affinity of phosphate groups with some metal ions or metal oxides. At present, the commonly used kits include the PhosphoProtein Purification kit and the ProteoEnrichTMATP-BindersTM kit. The latter is only used for the extraction and enrichment of protein kinase and other ATP-binding proteins (ABPs). After enrichment, the expression of phosphorylated proteins can be analyzed by means of two-dimensional electrophoresis and 2D-DIGE test.
This section mainly introduces the PhosphoProtein Purification kit as an example.
This experimental guide aims to help scientific researchers master the basic principle and main steps of affinity enrichment method for detecting protein phosphorylation level, and understand the application of affinity enrichment method for detecting phosphorylated protein.
Phosphoprotein Purification Kit is a system specially designed for phosphorylation of phosphorylated proteins in cell lysates. By using affinity chromatography to separate phosphorylated and non-phosphorylated proteins, we can obtain bioactive proteins, which are mainly used in proteomics, cell apoptosis and signal transduction. The principle of affinity chromatography is to use the principle of reversible combination of biopolymers and ligands to firmly bind the ligands to the carrier through covalent bonds to prepare a chromatographic system. The reversible binding mainly depends on the recognition of the ligand space structure of biopolymers.
After the completion of two-dimensional electrophoresis, carry out membrane transfer detection (isoelectric focusing electrophoresis in the first direction of two-dimensional electrophoresis, followed by SDS-PAGE) to separate proteins according to their relative molecular weight. The expression of phosphorylated protein was analyzed after Coomassie brilliant blue staining.
1. Main Instruments and Equipment
Micro-pipette, centrifuge, two-dimensional electrophoresis system, circulating water bath, decolorization shaking table, scanner, ImageMaster 2D Platinum version 5.0 software.
2. Material
Mouse liver nuclear cells.
3. Main reagents
(1) Electrophoretic buffer
(2) Colloidal reagent
(3) Stationary solution
(4) Coomassie brilliant blue dye solution
(5) Coomassie brilliant blue decoloring solution
(6) Hydration solution
(7) Balanced buffer
(8) Preparation of Bind-silane working solution
(9) Blocked agarose solution
(10) Tris-glycine electrophoresis buffer
(11) Enrichment kit (Phosphoprotein Purification kit)
(12) 2-D Clean-up kit
(13) 2-D Quant kit
1. Dissolution of nuclear cell protein
(1) Take a proper amount of nucleus, add the lysate, and dissolve it fully.
(2) 18000 g, centrifuged at 4°C for 35 min, and collected supernatant, which is nuclear protein.
2. Quantitative determination of nuclear cell protein
2-D Quant kit
3. Enrichment of phosphorylated protein
(1) Protein lysis
① Dilute an appropriate amount of protein sample with the lysis buffer provided by the 1500μL kit.
② Place it at 4°C for 30min and mix it every 10min.
③ 10000 g, centrifuged at 4°C for 30 min.
(2) Enrichment *1
In order to separate as many phosphorylated proteins as possible, we used 24 cm adhesive strips, pH 3-10, and made two parallel pieces of adhesive for each sample during electrophoresis, one for Kemas Brilliant Blue staining. One for semi-dry transfer and one for western blot detection.
① The purification column is prepared during centrifugation, and the column is washed with storage solution. Then balance the column with protein lysate containing 0.25% CHAPS for standby.
② Take the total protein supernatant containing 2.5 mg of protein, and adjust the concentration of protein lysate to 0.1 mg/mL to enrich phosphorylated protein. Finally, there will be 25mL of lysis products.

③ Take half the volume of pyrolysis liquid into the enrichment column.
④After all the pyrolysis liquid enters the rubber bed, add the remaining half of the pyrolysis liquid into the enrichment column.
⑤All liquids flow through the column, which takes about 50min. This step requires sufficient time to bind the phosphorylated protein to the affinity column.
⑥ The effluent was collected for detection of protein*2 that was not phosphorylated.
⑦ Clean the column with 6mL of cracking liquid.
(3) Collect phosphorylated protein
Use 500μL phosphorylated protein elution buffer is used to wash the phosphorylated protein from the column and repeat for 4 times for standby.
4. Quantification of phosphorylated proteins in nuclear cells
2-D Quant kit
5. Two-dimensional electrophoresis detection
In order to separate phosphorylated protein as much as possible, we use a 24cm adhesive strip, pH 3~11, and the sample loading amount is 150μg. Perform two-dimensional electrophoresis.
6. Coomassie brilliant blue staining (Figure 17-4-1)
A string of proteins with the same relative molecular weight and different isoelectric points (shown in the box) can be seen in the figure, suggesting that they may be the same protein with different phosphorylation levels.
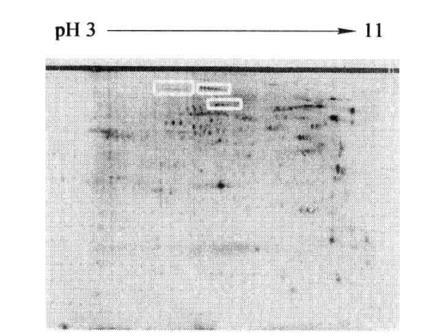
Figure 7-4-1 Two-dimensional electrophoresis map of phosphorylated protein in mouse liver nucleus
1. Obtaining enough high-purity total protein is very beneficial to the enrichment of phosphorylated protein.
2. When the kit is used for the first time, pay attention to prepare the preparation solution according to the instructions.
3. During the enrichment process, the column balance and other operations can be carried out on the ice, but after the protein sample is added to the column, all enrichment processes should be carried out at room temperature.
4. After obtaining rich phosphorylated protein, appropriate analysis methods should be selected. When the protein quality is large, ordinary two-dimensional electrophoresis analysis can be carried out, combined with coomassie brilliant blue staining, silver staining and other staining methods. When the protein quality is low, we can combine 2D-DIGE technology to analyze the differential expression of phosphorylated protein.
5. During elution, the eluent should be added in the middle of the column as much as possible, and should be placed for an appropriate time to fully elute the phosphorylated protein.
*1 The enrichment process should be carried out at 15~25°C, and the operation on ice is easy to reduce the enrichment efficiency.
*2 The effluent should be preserved, and then discarded after the phosphorylated protein is detected. If the detection results show that the phosphorylated protein is not enriched, it can be enriched again.

