Protocol of Preparation of Phosphorylated Protein Samples
The complexity and diversity of proteins in living organisms are not only reflected in the diversity of species but also in the structural variability. The response of organisms to the external environment can be regulated by conformational changes of proteins such as metastable effects and covalent modifications of protein primary structures, of which post-translational modifications are particularly important. Phosphorylation, glycosylation, and ubiquitination are three forms of modifications that are widespread in eukaryotic cells, and this section focuses on the enrichment of phosphorylation-modified proteins. It should be noted that, unlike the preparation of total and subcellular protein samples, the content of modified proteins is very low, so a simple enrichment followed by mass spectrometry is usually used to determine the modification sites and other information to avoid the loss of modified proteins and damage to the modification groups during the enrichment process.
To understand the types of biological protein modifications and their roles, and to master the basic strategies and main methods for enrichment of modified proteins. To gain a deep understanding of the importance of sample preparation for modified proteins.
Phosphorylation of proteins is one of the most important covalent modifications in organisms. Common separation and enrichment technologies for phosphorylated proteins/peptides include immobilized metal affinity chromatography (IMAC), metal oxide and metal hydroxide enrichment, antibody enrichment, strong cation exchange chromatography (SCX), strong anion exchange chromatography (SAX), etc. IMAC is the most commonly used method to separate phosphorylated peptides. In addition, because the phosphate group is easily removed by the phosphatase existing in the cell, it is very necessary to add a phosphatase inhibitor in the experiment. The separation and enrichment methods of phosphorylated proteins and peptides are shown in Table 1-3-1 and Table 1-3-2 respectively.
Table 1-3-1 Separation and Enrichment Methods of Phosphorylated Proteins
| Method | Antibody enrichment method | Kinase-specific enrichment | Affinity enrichment |
|---|---|---|---|
| Principle | Immunoprecipitation of target proteins from complex mixtures with specific antibodies that recognize phosphorylated amino acid residues. | Small molecule phosphokinase inhibitors were used for specific affinity chromatography. | Enrichment is carried out by the affinity between phosphoric acid group and some metal ions or metal oxides. |
Table 1-3-2 Separation and Enrichment Methods of Phosphorylated Peptides
| Method | Immobilized metal affinity chromatography, IMAC | Metal oxide and metal hydroxide enrichment method |
|---|---|---|
| Principle | Positively charged metal ions, such as Fe3+ and Ga3+, combine with negatively charged phosphate groups by electrostatic interaction. In the presence of high pH or phosphate, the binding between metal ions and phosphate groups is destroyed, and the phosphorylated peptide is released. | Enrichment of phosphorylated peptides by the affinity between phosphate groups and metal ions or metal oxides. TiO2 enrichment method is the most mature metal oxide enrichment method at present. |
| Method | Hydrophilic interaction chromatography, HILIC | Electrostatic repulsion-hydrophilic interaction chroma-tography, ERLIC |
| Principle | The separation is based on the hydrophilicity of the compound and the hydrogen bond and ion bond between the charged group and the stationary phase. Phosphorylated peptides are usually hydrophilic and charged due to their phosphate groups, and their binding ability in HILIC is stronger than that of non-phosphorylated peptides. The latest application of phosphorylated peptides is to separate them from non-phosphorylated peptides by hydrophilic interaction chromatography. | When pH ≤ 2, the N-terminal of the peptide segment carries a positive charge through the weak anion exchange column, which makes it have electrostatic repulsion with the chromatographic column packing. The phosphorylated peptide segment has electrostatic attraction with the packing due to the presence of negatively charged phosphate groups, which separates the phosphorylated peptide segment from the non-phosphorylated peptide segment. ERLIC is the latest method for the separation and enrichment of phosphorylated peptides. This method completes the enrichment and fractionation of peptides in one step experiment. |
| Method | Strong cation exchange chromatography, SCX | Strong anion exchange chromatography, SAX |
| Principle | The difference of charge between phosphorylated peptide and non-phosphorylated peptide in acid solution is used for separation. At pH2.7, most trypsin digestion products are charged with+2, while phosphorylated peptides are charged with -1 because they contain phosphate groups, so phosphorylated peptides are charged with +1 in acid solution. In strong cation exchange chromatography, the single charged peptide can flow out earlier than the multi charged peptide, and the phosphorylated peptide can be collected. | It is very similar to the basic principle of strong cation exchange chromatography. The strong acidity of the phosphate group on the phosphorylated peptide segment makes it easier to combine with the anion exchange chromatographic column, which has a strong retention capacity for the phosphorylated peptide segment, so as to achieve the purpose of separating and enriching the phosphorylated peptide segment. At present, SAX is often used in combination with other methods to obtain better enrichment effect of phosphorylated peptide, such as SAX and IMAC, SAX and SCX. |
| Method | MALDI target plate enrichment method | Chemical modification method |
| Principle | On line enrichment detection method combined with mass spectrometry identification. A small amount of simple sample is put on the target with special surface modification. After incubation, washing and elution, it is completely enriched. After mass spectrometry pretreatment, it is added with matrix for direct mass spectrometry analysis. | The phosphate group on the phosphorylated peptide was replaced by an affinity reagent, and the phosphorylated peptide was separated from the mixed peptide by affinity extraction. |
1. Main Instruments and Equipment
High-speed centrifuge, ultracentrifuge, ultrasonic crusher, vertical shaker, probe micro-ultrasonic crusher, PhosphoProtein Purification Column (product of Clontech), Nanosep ultrafiltration column (product of QIAGEN).
2. Experimental Materials
Protein sample of interest, usually a mixture of multiple proteins that have not been isolated.
3. Main reagents
(1) Lysis Solution
| Lysis solution A (applicable to the extraction of water-soluble protein) | |
| 40 mmol/L | Tris-base (pH 9. 5) |
| Lysis solution B (classic formula) | |
| 8 mol/L | Urea |
| 4% | CHAPS |
| 40 mmol/L | Tris-base |
| 1 % | DTT |
| 1 % | Protease inhibitor |
| Lysis solution based on classical formula (C - E) | |
| Lysis solution C | |
| 7 mol/L | Urea |
| 2 mol/L | Thiourea |
| 2 % - 4 % | CHAPS |
| 40 mmol/L | Tris-base |
| 1 % | DTT |
| 2 % | Pharmalyte (pH 3 - 10) |
| 1% | Protease inhibitor |
| Lysis solution D | |
| 9.5 mol/L | Urea |
| 2 % | CHAPS |
| 1 % | Tris-base |
| 0.8 % | Pharmalyte (pH 3 - 10) |
| 5 mmol/L | Protease inhibitor |
| Lysis solution E | |
| 9.5 mol/L | Urea |
| 2 % - 4 % | CHAPS |
| 1 % | DTT |
| 2 % | Pharmalyte (pH 3 - 10) |
| 5 mmol/L | Protease inhibitor |
| Lysis solution F (applicable to membrane protein extraction) | |
| 5 mol/L | Urea |
| 2 mol/L | Thiourea |
| 2 % | SB 3-10 |
| 2 % | CHAPS |
| 1 % | DTT |
| 0.5 % | CA |
| 5 mmol/L | Protease inhibitor |
| Lysis solution G (applicable to the extraction of insoluble precipitated protein) | |
| 1% | SDS |
| 0. 375 mol/L | Tris-HCl (pH 8.8) |
| 50 mmol/L | DTT |
| 25% (volume) | Glycerol |
| Lysis solution H(applicable to the PF-2D) | |
| 7.5 mol/L | Urea |
| 2.5 mol/L | Thiourea |
| 12.5 % | Glycerol |
| 62.5 mmol/L | Tris-HCl |
| 2. 5 % | n-Octyl-β-D-Glucopyranoside |
| 6.25 mmol/L | Tris (2-carboxyethyl) phosphine TCEP |
| 1.25 mmol/L | Protease inhibitor |
(2) Other Reagents
The following reagents are provided in the PhosphoProtein Purification Kit (Clontech product), which includes Phosphorylated Protein Lysate, Benzonase Reservoir*1, CHAPS, and Phosphorylated Protein Elution Solution.
(3) Protease Inhibitors
In the process of tissue cell fragmentation, the addition of diisopropyl fluorophosphate (DFP) can inhibit or slow down autolysis. The addition of iodoacetic acid can inhibit the activity of proteolytic enzymes that require sulfhydryl groups in active centers. The activity of proteolytic enzyme can also be eliminated by adding benzene sulfonyl fluoride (PMSF), which can be added according to the actual conditions in the experiment (Table 1-3-3). Use protease inhibitor complex tablets (Roche molecular Biochernicals), 5 mg/mL peptide inhibitor*2 prepared with 10 g/mL of DMSO as solvent.
Table 1-3-3 Broad-spectrum Protease Inhibitor Mixture
| Component | Final concentration |
|---|---|
| PMSF | 35 μg/mL (1 mmol/L) |
| EDTA | 0. 3 mg/mL (1 mmol/L) |
| Pepstatin | 0. 7 μg/mL |
| Leupeptin | 0. 5 μg/mL |
Enrichment Methods of Phosphorylated Proteins
(1) Configure phosphorylated protein lysate containing 0.25% CHAPS, 1 tablet of protease inhibitor complex and 10 μL of Benzonase stock solution and phosphorylated protein eluate containing 0.25% CHAPS.
(2) Add 5mL of phosphorylated protein lysate to the cultured cells, gently aspirate and beat, and resuspend the precipitate.
(3) Incubate at 4°C for 30min, with brief vortex shaking every 10min.
(4) Centrifuge at 10000g, 4°C for 30min*3.
(5) Collect the supernatant for quantification.
(6) Adjust the lysate containing approximately 2.5mg of total protein to a mass concentration of 0.1mg/mL with phosphorylated protein lysate to bring the total volume of lysate to 25mL*4.
(7) Add half of the lysate (12.5 mL) to the top of the phosphorylated protein purification column. Once almost all of the lysate is in the gel, add the other half of the lysate to the column and allow it to flow through the column. This step will take approximately 50 min and will require sufficient time for all phosphorylated proteins to be completely bound to the purification column*5*6.
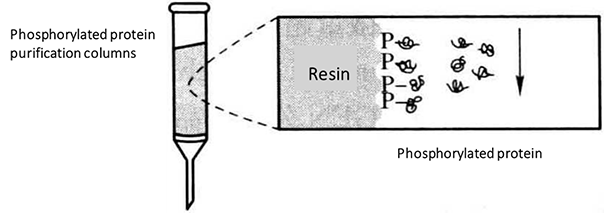
(8) Add 6 mL of phosphorylated protein lysate to the column and allow the liquid to flow through the column to clean the column.
(9) Add 500 μL of phosphorylated protein eluate containing 0.25% CHAPS to the column and collect the eluted components*7.
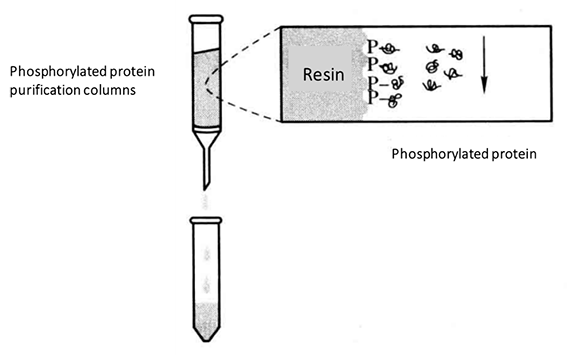
(10) Repeat step (9) four times to determine the concentration of all eluted components.
(11) Add 500 μL of protein sample to the Nanosep ultrafiltration column.
(12) Centrifuge at 10000g for 10min.
(13) After concentration, the sample that does not pass through the membrane can be aspirated back. It is recommended that the concentrated sample volume should not exceed 50 μL. If the sample is less than 50 μL, make up to 50 μL with buffer to obtain the desired yield.
(14) When the concentrated volume is greater than 500 μL, after the first centrifugation, remove the effluent, add diluted sample to the retained liquid to a total volume of 500 μL, and centrifuge again at 10,000g for 10 min.
* 1 Benzonase stock solution is a mixture of DNAase and RNAase.
* 2 Must be freshly prepared on the same day.
*3 Prepare the phosphorylated protein purification column during centrifugation. Remove the top and bottom caps of the column and drain the storage solution. Add 4 mL of phosphorylated protein lysate to allow the buffer to flow through to equilibrate the column.
*4 The increase in volume is to prevent complex formation during column passage for high protein concentrations, to maintain the solubilized state of the protein for full binding.
*5 Step (7) and the following steps must be performed at room temperature, never on ice, low temperatures will result in reduced binding.
*6 If analysis of unphosphorylated proteins in the lysate is required the fractions flowing through this step can be collected.
*7 The highest concentration of phosphorylated proteins in the eluted protein sample will be found in the 3rd and 4th elution fractions. After enrichment of the phosphorylated proteins, the protein components can be concentrated using Nanosep ultrafiltration columns.

