Sequence Based Protein Design
Sequence-Based Protein Design is a cutting-edge computational approach within the broader field of rational protein engineering. By analyzing the primary amino acid sequences of proteins, this method uncovers critical structural and functional insights encoded in the sequence itself. Through approaches such as multiple sequence alignment (MSA) and co-evolutionary analysis, researchers can identify conserved motifs, hotspot residues, and co-evolving amino acids that influence protein stability and functionality. Creative BioMart leverages these techniques alongside experimental validation to design proteins with enhanced properties, offering a comprehensive, one-stop service for rational protein design.
Understanding Sequence-Based Protein Design
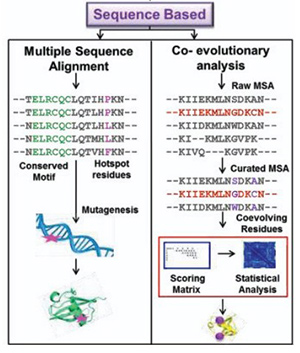
Figure 1. Sequence based computational approaches for designing and identifying different protein folds.(Gulati and Poluri, 2016)
Proteins derive their structural and functional characteristics largely from their amino acid sequences. The rational design of proteins, therefore, requires a detailed understanding of the information encoded in these sequences. Sequence-based protein design focuses on identifying residues critical for function, evolutionary adaptations, and mutational hotspots that may improve protein stability or activity. Two primary computational strategies dominate this approach:
- Multiple Sequence Alignments (MSA): MSA compares a target protein sequence with homologous sequences to identify conserved residues essential for structural integrity and functional activity. It allows researchers to pinpoint motifs, active sites, and regions suitable for mutagenesis. Widely used tools include Clustal Omega, MAFFT, K-Align, MUSCLE, and T-Coffee.
- Co-evolutionary Analysis: This method examines residue pairs that co-evolve within a protein family. Co-evolving residues often reflect functional dependencies or structural constraints, providing critical guidance for rational mutagenesis and protein optimization.
After computational predictions, candidate sequences undergo experimental validation to confirm their enhanced functionalities and stability. Techniques such as peptide synthesis, site-directed mutagenesis, or artificial gene synthesis—including incorporation of unnatural base pairs (UBPs)—enable researchers to translate in silico designs into functional proteins.
Our Services & Capacities
Creative BioMart provides professional sequence-based protein design services to support rational protein engineering projects. Our service portfolio includes:
-
Multiple Sequence Alignment (MSA)
Identify conserved motifs, active sites, and structurally essential residues using advanced alignment algorithms.
-
Co-evolutionary Analysis
Determine residue-residue correlations that inform rational mutagenesis and functional optimization.
-
Predicted Sequence Validation
Confirm design predictions through peptide synthesis, site-directed mutagenesis, or UBP-based gene construction.
-
Protein Expression & Purification
Produce the designed protein in relevant expression systems and purify it to homogeneity.
-
Functional and Structural Characterization
Evaluate activity, stability, and structural integrity using standard biochemical and biophysical assays.
Our integrated approach ensures that designed proteins not only exhibit theoretical improvements but are also experimentally validated for real-world applications.
Service Workflow

What We Need from You
To ensure accurate design and efficient project execution, clients are encouraged to provide:
- The target protein sequence(s) or accession number(s)
- Functional goals (e.g., increased activity, altered substrate specificity, improved solubility, or thermostability)
- Any available structural data, homology models, or literature references
- Expression preferences, such as host system or fusion tag requirements
- Desired experimental validations (computational only, or including lab-based testing)
What You Can Expect
Upon project completion, Creative BioMart delivers:
- A comprehensive sequence design report detailing alignment results, co-evolutionary analyses, and mutation rationales
- Optimized DNA or protein sequences ready for cloning or synthesis
- Optional expression constructs, purified proteins, or analytical data packages
- Experimental validation results confirming predicted enhancements (where applicable)
- Full technical support and consultation to interpret findings and plan next-stage development
What Sets Us Apart
- Expertise in Rational Design: Decades of experience in computational and experimental protein engineering.
- Comprehensive Workflow: From sequence analysis to experimental validation and protein characterization.
- Cutting-Edge Tools: Use of industry-standard software for MSA and co-evolutionary analysis.
- Custom Solutions: Designs tailored to your protein of interest and desired properties.
- Integrated Experimental Validation: Ensures theoretical designs are functionally robust.
- Timely and Reliable Delivery: Structured workflow guarantees project efficiency and reproducibility.
Case Studies and Real-World Applications
Case 1: Automated sequence-based design of proteins for high bacterial expression and stability
Goldenzweig et al., 2016. doi:10.1016/j.molcel.2016.06.012
Sequence-based protein design offers a powerful computational approach to enhance protein stability and expression without compromising function. Using a fully automated, web-based algorithm, researchers successfully stabilized five challenging proteins, including human acetylcholinesterase (hAChE). A designed hAChE variant with 51 mutations exhibited a ~2,000-fold increase in soluble expression in E. coli and a 20°C rise in thermostability while maintaining native activity and structure. The method improves core packing, surface polarity, and backbone rigidity using only a 3D structure and homologous sequences, demonstrating exceptional versatility and efficiency for designing stable, high-yield protein variants across diverse species.
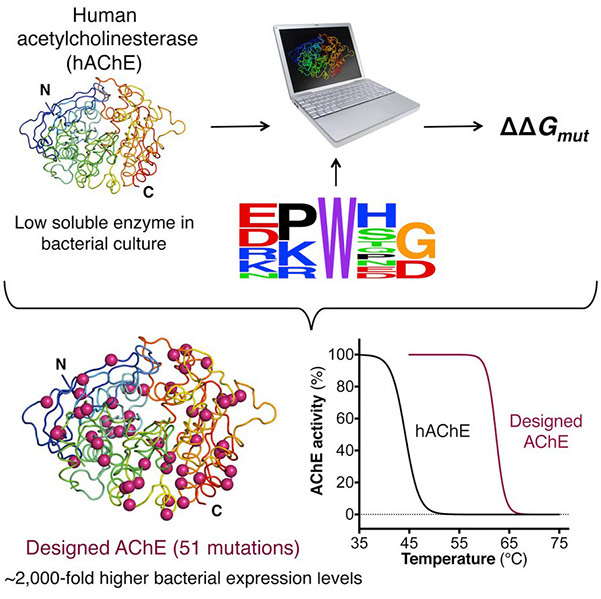
Figure 2. A new computational method is used to stabilize five recalcitrant proteins. The designed variants show higher expression and stability with unmodified function. (Goldenzweig et al., 2016)
Case 2: Co-evolutionary analysis into Domain–Domain Interactions
Jothi et al., 2006. doi:10.1016/j.jmb.2006.07.072
Recent advances in functional genomics have enabled large-scale mapping of protein–protein interactions; however, these data often lack information about the specific domains mediating such interactions. To address this, researchers performed co-evolutionary and structural analyses across protein complexes such as F1-ATPase, Sec23p/Sec24p, RNA polymerase, and nuclear pore complexes, revealing that interacting domain pairs exhibit stronger co-evolution than non-interacting ones. Building on this observation, a computational method was developed to predict domain–domain interactions genome-wide. Applied to the yeast interactome and validated using PDB structures, the method demonstrated statistically significant accuracy and uncovered previously undetected interaction sites, complementing existing prediction approaches.
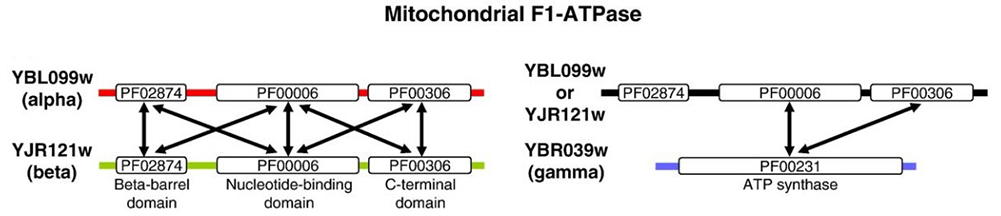
Figure 3. Protein sequences are shown using thick colored lines: red for the alpha chain, green for the beta chain, blue for the gamma chain, and black for alpha or beta chain. Pfam domain annotations are shown using rectangular boxes (not drawn to scale). The names of the protein sequences are to the left of the domain architecture. Inter-chain domain–domain interactions, which are known to be true from PDB crystal 0structures (as inferred in iPfam), are shown using double-arrow lines in the domain architecture. (Jothi et al., 2006)
Customer Testimonials: Success Stories with Sequence-Based Design
"Creative BioMart’s sequence-based design expertise was instrumental in optimizing a therapeutic enzyme for improved thermal stability. Their team performed an in-depth co-evolutionary analysis that pinpointed mutational hotspots, and the resulting variant retained full catalytic activity while showing a 3-fold increase in half-life under industrial conditions. The seamless integration of computational predictions with experimental validation impressed our internal team."
— Senior Scientist | Biotech Enzyme Company
"We engaged Creative BioMart to design an antibody fragment with enhanced binding affinity. Their multiple sequence alignment and hotspot residue analysis provided a clear roadmap for mutagenesis. The experimental validation and subsequent protein expression delivered a variant with significantly improved antigen recognition and high solubility, accelerating our preclinical program."
— Director of R&D | Mid-Sized Biopharmaceutical Company
"For a synthetic protein scaffold project, Creative BioMart applied advanced sequence-based design coupled with UBP-enabled gene synthesis. The team guided us through computational optimization, gene construction, and functional characterization. The resulting scaffold exhibited remarkable folding stability and retained the desired functional motifs, which was critical for our downstream protein engineering applications."
— Head of Protein Engineering | Synthetic Biology Startup
"Creative BioMart helped us redesign a metabolic enzyme to improve both activity and stability. Their sequence-based approach identified conserved motifs and co-evolving residues, which informed targeted mutagenesis. After expression and characterization, the redesigned enzyme exceeded our activity benchmarks and demonstrated superior stability under process-relevant conditions, shortening our development timeline considerably."
— R&D Director | Industrial Biotechnology Company
FAQs About Sequence-Based Protein Design
-
Q: What is sequence-based protein design and how does it differ from other protein engineering approaches?
A: Sequence-based protein design is a computational approach that focuses on the amino acid sequence of a protein to identify critical residues, evolutionary patterns, and mutational hotspots. Unlike structure-based approaches, which rely heavily on 3D protein models, sequence-based design leverages evolutionary information and conserved motifs to guide rational modifications, making it particularly powerful for improving stability, activity, or specificity even when structural data is limited. -
Q: What types of proteins can be optimized using this service?
A: Our service is highly versatile and can be applied to a wide range of proteins, including therapeutic antibodies, enzymes, synthetic scaffolds, and metabolic proteins. Whether your goal is to improve stability, activity, binding affinity, or solubility, our computational and experimental workflows can be tailored to your protein of interest. -
Q: What computational tools do you use for sequence analysis?
A: We utilize industry-standard tools for multiple sequence alignment (MSA) such as Clustal Omega, MAFFT, MUSCLE, K-Align, and T-Coffee. For co-evolutionary analysis, we apply algorithms that detect correlated mutations and critical residue pairs, providing insights into functional dependencies and structural constraints. These analyses form the basis for rational protein design. -
Q: How do you validate the designed protein sequences?
A: Designed sequences undergo thorough experimental validation. Depending on the project, we use peptide synthesis, site-directed mutagenesis, or artificial gene synthesis with unnatural base pairs (UBPs). Validated sequences are then expressed, purified, and characterized to confirm improvements in stability, functionality, and expression performance. -
Q: Can you assist with protein expression and purification after design?
A: Absolutely. We provide end-to-end support, from computational design to expression, purification, and characterization of your target protein. This ensures that the theoretical improvements suggested by our sequence-based analysis translate into practical, functional proteins ready for downstream applications. -
Q: How long does a typical project take?
A: Project timelines vary depending on the protein type, design complexity, and validation methods. Typically, sequence analysis and design take a few weeks, followed by experimental validation and protein production. We provide a customized timeline during project consultation to ensure realistic and efficient project planning.
Other Resources
Related Services
- Protein Engineering Services
- Structure Based Protein Design
- De Novo Protein Design
- Protein Expression and Purification Services
- Protein Analytical Service
- Computer-aided Enzyme Design
- Site-Directed Mutagenesis
- Protein Sequence Analysis and Function Prediction
Related Products
References:
- Goldenzweig A, Goldsmith M, Hill SE, et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Molecular Cell. 2016;63(2):337-346. doi:10.1016/j.molcel.2016.06.012
- Gulati K, M. Poluri K. An overview of computational and experimental methods for designing novel proteins. BIOT. 2016;10(3):235-263. doi:10.2174/1872208310666161013152249
- Jothi R, Cherukuri PF, Tasneem A, Przytycka TM. Co-evolutionary analysis of domains in interacting proteins reveals insights into domain–domain interactions mediating protein–protein interactions. Journal of Molecular Biology. 2006;362(4):861-875. doi:10.1016/j.jmb.2006.07.072
- Wang J. Protein sequence design by deep learning. Nat Comput Sci. 2022;2(7):416-417. doi:10.1038/s43588-022-00274-5
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

