Special Cell-based Display Platform
Creative BioMart’s Special Cell-based Display Platform harnesses the power of Gram-positive bacteria, including Bacillus subtilis, lactic acid bacteria (LAB), and Corynebacterium glutamicum, to enable protein engineering under unique and harsh conditions. By displaying proteins on cell surfaces or stress-resistant endospores, this platform allows directed evolution for extreme stability, catalytic efficiency, or binding properties unattainable with conventional systems. Combined with high-throughput screening, clone recovery, and recombinant protein expression, our platform provides a comprehensive, one-stop solution for the discovery and optimization of proteins with specialized functions for industrial, therapeutic, or research applications.
Understanding Yeast Display Technology
While E. coli, yeast, and phage display systems dominate protein engineering, Gram-positive bacteria and spore-based systems offer unique advantages. The rigid cell walls of Gram-positive bacteria make them ideal for whole-cell catalysts and adsorbents, and their endospores provide unmatched resistance to extreme chemical and physical conditions. Display strategies include:
- Noncovalent attachment to cell wall peptidoglycan via LysM domains.
- Covalent anchoring using LPXTG motifs processed by sortase transpeptidase.
- Spore display, fusing target proteins to coat proteins such as CotB, CotC, or CotG, allowing folding-free expression and functional protein display under harsh environments.
These specialized systems enable the evolution of proteins with enhanced stability, resistance, and extreme functional properties, expanding the range of applications beyond conventional hosts.
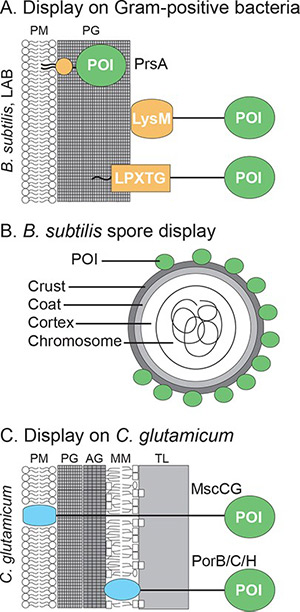
Figure 1. Surface display systems developed in Gram-positive bacteria. (Huang and Clubb, 2017)
What Our Special Cell-based Display Platform Deliver
Creative BioMart provides a full suite of services for Gram-positive and spore-based protein display:
- Display of target proteins on Gram-positive bacterial surfaces (e.g., B. subtilis, LAB, C. glutamicum )
- Construction of mutagenic and combinatorial libraries for functional diversification
- High-throughput screening under standard or extreme conditions to identify superior variants
- Recovery and sequencing of selected clones
- Recombinant protein expression, purification, and functional characterization
- Tailored strategies for enzymes, binding proteins, and extreme-condition functional proteins
Service Workflow

Service Features
|
Features |
Details |
|---|---|
|
Library Diversity |
Up to 10⁸–10¹⁰ unique sequences. |
|
Display Systems |
Surface display using LysM domains or LPXTG motifs; spore-based display using CotB, CotC, or CotG. |
|
Screening Capabilities |
High-throughput assays including conditions mimicking industrial or extreme environments. |
|
Protein Types |
Enzymes, binding proteins, industrial biocatalysts, and stress-resistant variants. |
|
Applications |
Biocatalysis, adsorbents, therapeutic protein stabilization, industrial enzymes, extreme-condition evolution. |
Our Distinguishing Advantages
- Validated Gram-positive Display Platform: Optimized for B. subtilis and other strains.
- High Library Diversity: Supports generation of up to 10¹⁰ variants for thorough exploration.
- Screening Under Harsh Conditions: Enables evolution of proteins for extreme stability or activity.
- One-Stop Protein Engineering: From display design to recombinant protein characterization.
- Expertise in Spore-Based Systems: Unique capabilities for endospore-displayed proteins.
- Tailored Solutions: Customizable strategies for enzymes, therapeutic proteins, and industrial targets.
Case Studies of Protein Engineering Using a Special Cell-based Display Platform
Case 1: Spore-based display of active feed enzymes
Potot et al., 2010. doi:10.1128/AEM.01103-10
The use of bacterial spore systems to display feed enzymes offers a promising approach to enhance nutrient digestibility in animal feed while reducing environmental impact. Spores’ natural resistance to harsh feed preparation and gastrointestinal conditions enables enzymes to remain active during delivery. In this study, the inner-coat protein OxdD of Bacillus subtilis was successfully used to display active phytase on the spore surface, outperforming the more abundant CotG outer-coat protein. Additionally, OxdD effectively displayed a bioactive heterologous enzyme, β-glucuronidase, demonstrating its potential as a versatile carrier for functional enzyme display in feed and other applications.
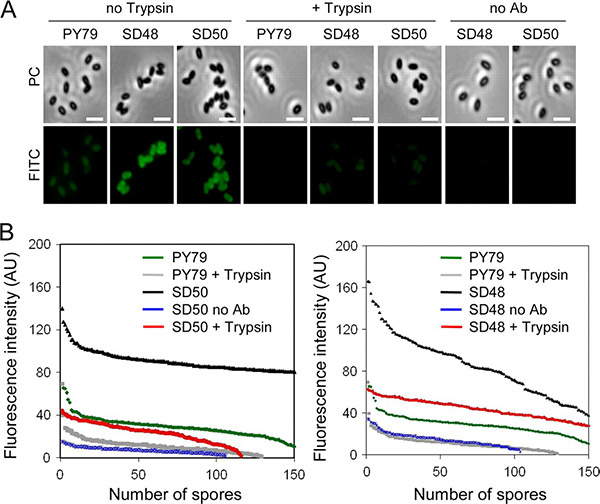
Figure 2. Immunofluorescence detection of CotG-Phy and OxdD-Phy protein fusions at the surface of purified spores. FITC, fluorescein isothiocyanate. (A) Phase-contrast microscopy (PC) and fluorescence microscopy (FITC). Scale bars, 2 μm. (B) Fluorescence intensity was quantified on 150 spores. AU, arbitrary units. noAb, control without primary antibody. Spores were purified from cultures of the following strains: PY79, wild type; SD48, CotG-Phy; and SD50, OxdD-Phy. (Potot et al., 2010)
Case 2: Heterologous protein display on the cell surface of lactic acid bacteria mediated by the S-layer protein
Hu et al., 2011. doi:10.1186/1475-2859-10-86
Researchers developed a novel surface display system for lactic acid bacteria (LAB) using the C-terminal region (LcsB) of the Lactobacillus crispatus S-layer protein SlpB, responsible for cell wall anchoring. Sequence analysis showed LcsB’s strong similarity to known cell wall-binding domains. When fused with green fluorescent protein (GFP) or β-galactosidase (Gal), these fusion proteins bound effectively to various LAB surfaces, with improved binding after SDS pretreatment. Both proteins could co-display on the same cell, and GFP was successfully expressed on Lactococcus lactis surfaces using LcsB as an anchor. These findings demonstrate LcsB’s potential as a versatile scaffold for biotechnological and functional display applications.
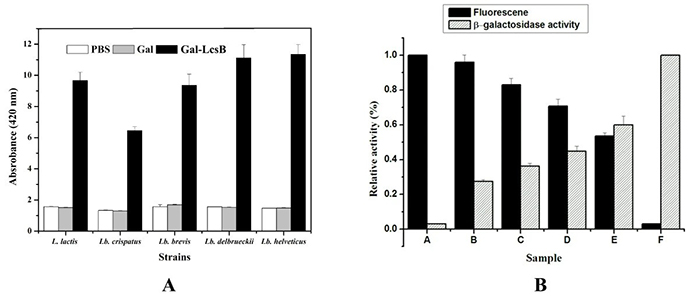
Figure 3. Functional display of β-galactosidase and dual-protein presentation on LAB cells. (A) Surface β-galactosidase activity of various LAB strains after incubation with PBS, Gal, or Gal-LcsB. (B) Co-display of GFP-LcsB and Gal-LcsB on L. brevis cells at different ratios, showing corresponding fluorescence intensity and enzyme activity (A–F represent increasing proportions of Gal-LcsB). (Hu et al., 2011)
Customer Experiences with Our Special Cell-based Display Platform
“We partnered with Creative BioMart to evolve a thermostable cellulase using their Bacillus subtilis spore display platform. Their team designed the library and high-throughput screening process, and the resulting variants retained activity at extreme temperatures and pH levels. This capability would have been impossible with conventional E. coli or yeast systems. Their expertise and streamlined workflow greatly accelerated our industrial enzyme development program.”
— Senior Scientist | Industrial Biotechnology Company
“Using Creative BioMart’s Gram-positive bacterial display, we engineered a whole-cell catalyst for biomass degradation. The team optimized protein surface display and high-throughput screening, producing strains with significantly enhanced catalytic efficiency. The robustness of the system allowed us to screen under conditions mimicking industrial fermentation, saving both time and cost.”
— Head of Enzyme R&D | Biofuel Manufacturer
“Creative BioMart helped us develop stress-resistant enzyme variants using their endospore display system. The protein folding challenges that typically arise in bacterial expression were eliminated, and the clones exhibited remarkable stability in harsh chemical environments. Their integrated approach from library design to recombinant protein characterization was invaluable.”
— Director of Protein Engineering | Specialty Chemicals Company
“We used Creative BioMart’s special cell-based display platform to evolve binding proteins for industrial adsorption applications. The combination of Gram-positive surface display and high-throughput selection under extreme conditions allowed us to identify clones with superior binding affinity and durability. Their expertise and attention to detail were outstanding.”
— Principal Investigator | Environmental Biotechnology Start-up
FAQs About Our Special Cell-based Display Platform
-
Q: Which Gram-positive bacteria are supported for display?
A: We support a range of Gram-positive strains, including Bacillus subtilis, lactic acid bacteria (LAB), and Corynebacterium glutamicum, depending on the application and desired protein display system. -
Q: Can this platform be used for enzymes that need extreme stability?
A: Yes. Our spore-based display system allows proteins to be evolved and screened under harsh chemical, thermal, and oxidative conditions, making it ideal for industrial enzyme optimization. -
Q: How is protein displayed on Gram-positive cells?
A: Proteins can be anchored to the cell wall via noncovalent LysM domains, covalent LPXTG motifs processed by sortase, or fused to endospore coat proteins (CotB, CotC, CotG) for stress-resistant display. -
Q: What is the typical library size achievable?
A: Libraries can reach up to 10⁸–10¹⁰ unique variants, allowing comprehensive exploration of sequence space for functional improvements. -
Q: Can selected clones be expressed recombinantly?
A: Yes. Clones recovered from the display system can be transferred to expression systems for recombinant protein production, purification, and functional testing. -
Q: What applications are best suited for this platform?
A: Ideal applications include whole-cell catalysts, adsorbents, industrial enzymes, stress-resistant proteins, and any protein requiring evolution under extreme chemical, physical, or thermal conditions.
Other Resources
Related Services
- Protein Engineering Services
- Directed Evolution
- Protein Expression and Purification Services
- Phage Display Platform
- Escherichia coli Display Platform
- Yeast Display Platform
- Cell-free Display Platform
Related Products
References:
- Hu S, Kong J, Sun Z, Han L, Kong W, Yang P. Heterologous protein display on the cell surface of lactic acid bacteria mediated by the S-layer protein. Microb Cell Fact. 2011;10(1):86. doi:10.1186/1475-2859-10-86
- Huang GL, Clubb RT. Progress towards engineering microbial surfaces to degrade biomass. In: Tumuluru JS, ed. Biomass Volume Estimation and Valorization for Energy. InTech; 2017. doi:10.5772/65509
- Potot S, Serra CR, Henriques AO, Schyns G. Display of recombinant proteins on Bacillus subtilis spores, using a coat-associated enzyme as the carrier. Appl Environ Microbiol. 2010;76(17):5926-5933. doi:10.1128/AEM.01103-10
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

