Structure Based Protein Design
Sequence-Based Protein Design is a cutting-edge computational approach within the broader field of rational protein engineering. By analyzing the primary amino acid sequences of proteins, this method uncovers critical structural and functional insights encoded in the sequence itself. Through approaches such as multiple sequence alignment (MSA) and co-evolutionary analysis, researchers can identify conserved motifs, hotspot residues, and co-evolving amino acids that influence protein stability and functionality. Creative BioMart leverages these techniques alongside experimental validation to design proteins with enhanced properties, offering a comprehensive, one-stop service for rational protein design.
Understanding Structure-Based Protein Design
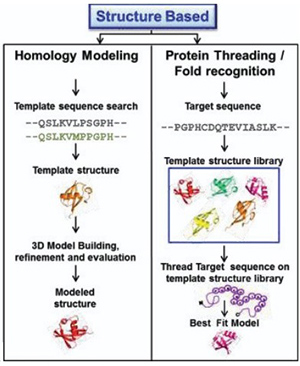
Figure 1. Sequence based computational approaches for designing and identifying different protein folds.(Gulati and Poluri, 2016)
Proteins function through complex three-dimensional structures stabilized by intricate networks of non-covalent interactions. Understanding and manipulating these structural features are at the heart of rational protein design. Structure-Based Protein Design (SBPD) employs computational modeling techniques to predict, analyze, and redesign protein structures for improved properties such as enhanced catalytic efficiency, increased thermostability, or altered substrate specificity.
Several established computational approaches fall under the umbrella of SBPD:
- Homology Modeling: Relies on structural similarity between homologous proteins. If a template structure of a closely related protein is available, this method predicts the 3D structure of the target protein with high accuracy.
- Protein Threading (Fold Recognition): Suitable for cases where no close homologs exist; it predicts the most probable fold of a sequence by matching it to known structural frameworks.
- Fragment-Based Methods: Utilize known structural fragments from databases to assemble partial models when no complete template is available.
- Ab Initio Methods: Employ all-atom simulations based on potential energy functions, requiring no template structures, to predict protein conformation from scratch.
Following computational design, experimental validation—via peptide synthesis, site-directed mutagenesis, or gene synthesis incorporating unnatural base pairs (UBPs)—ensures that designed variants perform as predicted in real biological systems.
Structure Based Protein Design: Services & Capacities
Creative BioMart delivers expert services in structure-based protein design, combining advanced computational modeling with rigorous experimental validation. Our offerings include:
-
Homology Modeling
Leverages known structural templates to predict 3D structures and identify functional regions for rational engineering.
-
Protein Threading
Aligns target sequences to existing fold libraries to predict the most compatible 3D framework.
-
Fragment-Based Methods
Assemble partial structures when full-length templates are unavailable, enabling the modeling of complex or multi-domain proteins.
-
Ab Initio Simulation
Conducts all-atom modeling using energy minimization techniques to predict novel folds independent of known structures.
-
Experimental Validation
Employs site-directed mutagenesis, synthetic gene construction, and expression analysis to confirm the functional success of designed proteins.
-
Characterization
Includes biophysical and biochemical assays to assess folding, stability, and activity.
Our integrated workflow ensures each project moves seamlessly from computational prediction to experimental verification.
Service Workflow

Requirements and Deliverables
-
Information We Require from Clients
To ensure accurate modeling and efficient project progression, clients are encouraged to provide:
- Target protein sequence(s) or corresponding UniProt/PDB identifiers
- Any available structural data, such as crystal or cryo-EM structures, or homology templates
- Functional objectives, such as enhancing stability, modifying binding affinity, or improving expression yield
- Preferred expression systems (e.g., E. coli, yeast, insect, or mammalian cells)
- Experimental preferences, including desired assays or validation depth ( in silico only, or computational + experimental)
-
Results Clients Can Receive
At the completion of a project, Creative BioMart provides:
- A comprehensive modeling report detailing methodology, templates used, structural quality metrics, and mutation rationale
- 3D structural models in standard file formats (e.g., PDB, mmCIF) for visualization or docking studies
- Designed sequences optimized for expression and functionality
- Optional validated experimental data, including protein expression, purification, and characterization results
Why Choose Creative BioMart
- Expertise in Rational Design: Decades of experience in computational and experimental protein engineering.
- Comprehensive Workflow: From sequence analysis to experimental validation and protein characterization.
- Cutting-Edge Tools: Use of industry-standard software for MSA and co-evolutionary analysis.
- Custom Solutions: Designs tailored to your protein of interest and desired properties.
- Integrated Experimental Validation: Ensures theoretical designs are functionally robust.
- Timely and Reliable Delivery: Structured workflow guarantees project efficiency and reproducibility.
Case Studies and Real-World Applications
Case 1: Structure-based redesign of enzymes for fluoroquinolone biodegradation
Liu et al., 2019. doi:10.3390/ijerph16183407
To enhance the biodegradation efficiency of fluoroquinolone antibiotics during sewage treatment, researchers applied structure-based protein design using molecular docking and homology modeling. Binding site residues of aerobic, anaerobic, and facultative degrading enzymes were analyzed, and hydrophobic residues near the fluoroquinolone molecules were replaced with hydrophilic ones to improve binding. A total of 150 redesigned enzyme variants were modeled and evaluated for binding affinity to ciprofloxacin, norfloxacin, and ofloxacin. Several modified enzymes showed over 50% increased degradation efficiency, with one anaerobic variant achieving affinity improvements exceeding 160%. Energy calculations confirmed reduced reaction barriers, validating the enhanced catalytic degradation performance.
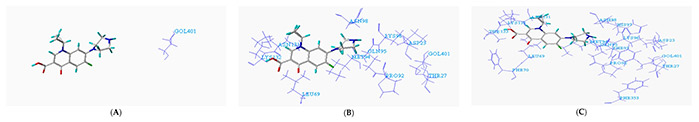
Figure 2. Conformation of ciprofloxacin (CIP) and degrading enzyme (PDB ID: 1YZP). (A) Within 1Å (GOL401); (B) Within 2Å (ASP23, THR27, LEU69, PRO92, GLN95, LYS96, ASN98, ASN131, LYS132, GOL401); (C) Within 5Å (ASP23, THR27, LEU69, PHE70, PRO92, MET94, GLN95, LYS96, HIS97, ASN98, ASN131, LYS132, THR133, PHE353). (Liu et al., 2019)
Case 2: Fragment-based ligand design of novel potent inhibitors of tankyrases
Larsson et al., 2013. doi:10.1021/jm400211f
Tankyrases are promising therapeutic targets for cancer and myelin-degrading diseases. Using a structure- and biophysics-driven fragment-based ligand design strategy, researchers identified a novel class of potent tankyrase inhibitors. Thermal shift assays pinpointed efficient fragments binding at the nicotinamide site—a key hotspot for ligand interaction. Optimization of the initial fragment, 4-methyl-1,2-dihydroquinolin-2-one, significantly enhanced binding affinity. Crystal structure analysis of an optimized analog revealed unique interactions exploiting structural distinctions within the tankyrase active site. Further refinement produced inhibitors with low nanomolar IC50 values, excellent solubility, PARP selectivity, and high ligand efficiency, demonstrating the power of structure-based protein design in drug discovery.
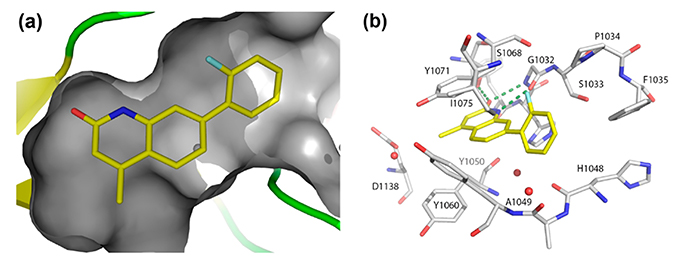
Figure 3. Surface representation showing the overall positioning of compound 11 in the pocket. (b) Crystal structure of the active site of TNKS2 in complex with 11 showing the nonplanar binding conformation of the o-fluorophenyl moiety with hydrophobic interaction with Ile1075, Phe1035, and Tyr1050 as well as the o-fluoro interaction with the main chain oxygen of Tyr1071. (Larsson et al., 2013)
What Our Clients Say About Our Structure-Based Protein Design Service
"Creative BioMart’s structure-based protein design service was pivotal in optimizing our enzyme for industrial biocatalysis. Their team used homology modeling and molecular dynamics simulations to identify key residues influencing stability. The redesigned variant exhibited a 25°C increase in thermostability and retained over 95% of its catalytic efficiency. Their precise modeling and experimental validation shortened our development timeline considerably."
— Director of Bioprocess Development | Industrial Biotechnology Company
"We collaborated with Creative BioMart to enhance the binding affinity of a therapeutic antibody fragment. Through structure-based modeling and energy minimization, they pinpointed amino acid substitutions in the complementarity-determining regions. The resulting variant showed a fourfold improvement in antigen affinity, confirmed through surface plasmon resonance assays. Their scientific rigor and responsiveness made the entire process seamless."
— Head of Therapeutic Antibody R&D | Mid-Sized Biopharmaceutical Company
"Our team approached Creative BioMart to model and stabilize a membrane receptor that was difficult to express. Using protein threading and fragment-based modeling, their scientists successfully generated a high-confidence 3D structure, allowing targeted redesign of flexible regions. The optimized construct expressed robustly in E. coli and yielded diffraction-quality crystals. Their technical expertise exceeded our expectations."
— Principal Investigator | Academic Structural Biology Laboratory
"Creative BioMart helped us redesign a key metabolic enzyme in our synthetic biology pipeline. Their ab initio modeling and mutational analysis guided us toward structural improvements that enhanced solubility and reduced aggregation during fermentation. The final construct expressed efficiently and maintained native function. Their integrated computational-experimental workflow delivered both scientific insight and tangible performance gains."
— R&D Director | Synthetic Biology Startup
FAQs About Structure-Based Design
-
Q: What is Structure-Based Protein Design, and how does it differ from Sequence-Based Protein Design?
A: Structure-Based Protein Design (SBPD) focuses on the three-dimensional (3D) structure of a protein to understand and engineer its function. By analyzing non-covalent interactions, folding patterns, and active sites, SBPD enables atomic-level modification of protein properties. In contrast, Sequence-Based Protein Design relies on amino acid sequence data and evolutionary patterns. While sequence-based approaches are powerful when structures are unknown, structure-based design provides more precise, targeted control when 3D structural information is available. -
Q: What types of proteins can benefit from structure-based design?
A: SBPD is suitable for a wide range of proteins, including enzymes, receptors, antibodies, transporters, and synthetic scaffolds. It is especially effective for improving stability, catalytic activity, substrate specificity, binding affinity, or folding efficiency. Whether for industrial enzymes, therapeutic proteins, or research reagents, our design strategies can be customized to meet specific functional or stability goals. -
Q: Do I need to provide a known 3D structure for my target protein?
A: Not necessarily. If a high-resolution structure is available, it significantly enhances precision. However, when no structure exists, we can construct reliable models using homology modeling, threading, or ab initio methods. Our modeling expertise ensures that even without a known structure, we can predict and optimize your protein’s 3D conformation accurately. -
Q: How do you validate the designed proteins experimentally?
A: After computational design, we perform experimental validation using a combination of peptide synthesis, site-directed mutagenesis, or artificial gene synthesis, including systems incorporating unnatural base pairs (UBPs). Designed variants are then expressed, purified, and characterized to verify their folding, stability, and functional activity—ensuring that computational predictions translate effectively into experimental success. -
Q: How long does a structure-based protein design project typically take?
A: Project duration depends on protein complexity, design objectives, and the chosen validation methods. Typically, computational modeling and in silico optimization require several weeks, while experimental validation and protein characterization may take additional time. We provide a detailed project timeline after initial consultation, ensuring transparent expectations and efficient progress. -
Q: Can this service be combined with sequence-based protein design?
A: Absolutely. Many clients choose to integrate sequence-based and structure-based design strategies for optimal results. Sequence analysis helps identify conserved residues and mutational hotspots, while structure-based modeling provides spatial context for those changes. This hybrid approach enhances both accuracy and success rates in achieving desired protein properties. -
Q: What industries or research areas can benefit most from your SBPD services?
A: Our structure-based protein design solutions are valuable across pharmaceuticals, biotechnology, enzyme engineering, synthetic biology, and academic research. Applications include developing more stable industrial enzymes, improving therapeutic protein formulations, designing novel protein scaffolds, and creating customized biomolecules for diagnostics or biosensors.
Other Resources
Related Services
- Protein Engineering Services
- Sequence Based Protein Design
- De Novo Protein Design
- Protein Expression and Purification Services
- Protein Analytical Service
- Computer-aided Enzyme Design
Related Products
References:
- Gulati K, M. Poluri K. An overview of computational and experimental methods for designing novel proteins. BIOT. 2016;10(3):235-263. doi:10.2174/1872208310666161013152249
- Larsson EA, Jansson A, Ng FM, et al. Fragment-based ligand design of novel potent inhibitors of tankyrases. J Med Chem. 2013;56(11):4497-4508. doi:10.1021/jm400211f
- Liu S Cheng, Sun S Jun, Cui P, Ding Y Fan. Molecular modification of fluoroquinolone-biodegrading enzymes based on molecular docking and homology modelling. IJERPH. 2019;16(18):3407. doi:10.3390/ijerph16183407
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

