PDE6C
-
Official Full Name
PDE6C -
Overview
Cone cGMP-specific 3', 5'-cyclic phosphodiesterase subunit alpha' is an enzyme that in humans is encoded by the PDE6C gene. -
Synonyms
5-cyclic phosphodiesterase subunit alpha;cGMP phosphodiesterase 6C;COD4;Cone cGMP specific 3 5 cyclic phosphodiesterase subunit alpha;Cone cGMP-specific 3;PDE 6C;PDE6 alpha prime;PDE6 alpha;PDE6C;PDEA2;Phosphodiesterase 6C cGMP specific cone alpha prime
| Cat.# | Product name | Source (Host) | Species | Tag | Protein Length | Price |
|---|---|---|---|---|---|---|
| PDE6C-11119Z | Recombinant Zebrafish PDE6C | Mammalian Cells | Zebrafish | His |
|
|
| PDE6C-12560M | Recombinant Mouse PDE6C Protein | Mammalian Cells | Mouse | His |
|
|
| PDE6C-444H | Recombinant Human PDE6C protein, GST-tagged | Insect Cells | Human | GST |
|
|
| PDE6C-6544C | Recombinant Chicken PDE6C | Mammalian Cells | Chicken | His |
|
|
| PDE6C-6589M | Recombinant Mouse PDE6C Protein, His (Fc)-Avi-tagged | HEK293 | Mouse | Avi&Fc&His |
|
|
| PDE6C-6589M-B | Recombinant Mouse PDE6C Protein Pre-coupled Magnetic Beads | HEK293 | Mouse |
|
Background
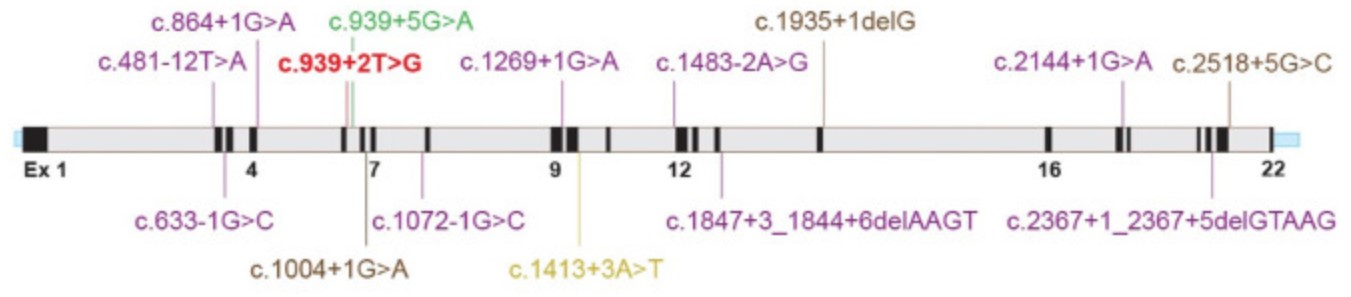
Fig1. Illustration of PDE6C gene (NM_006204.4) with novel splicing (this study) and reported splicing/small deletion variants (HGMD).
What is PDE6C protein?
PDE6C (Phosphodiesterase 6C) gene is a protein coding gene. The PDE6C gene is situated on the long arm of chromosome 10 at locus 10q.23 that spans approximately 53.4 kb of genomic DNA.
PDE6C gene encodes the alpha-prime subunit of cone phosphodiesterase, which is composed of a homodimer of two alpha-prime subunits and 3 smaller proteins of 11, 13, and 15 kDa. The PDE6C protein is consisted of 858 amino acids and its molecular mass is approximately 99 kDa. The subcellular locations for PDE6C is in cell membrane lipid-anchor and cytoplasmic side.
Phosphodiesterases (PDEs) are a family of phosphohydrolyases that catalyze the hydrolysis of 3' cyclic phosphate bonds in adenosine and/or guanine 3',5' cyclic monophosphate (cAMP and/or cGMP). PDEs regulate the second messengers by controlling their degradation. PDE6 is an essential component of the phototransduction cascade, which converts light to electrical signals within the neural retina. Rods have a PDE6 catalytic core composed of a heterodimer of PDE6A and PDE6B subunits (inhibited by PDE6G), whereas cones have a catalytic homodimer of PDE6C (inhibited by PDE6H subunits). These three proteins are more closely related to each other than to the remaining phosphodiesterases, and available frog and fish sequences support early vertebrate origin.
What is the function of PDE6C protein?
As cone-specific cGMP phosphodiesterase, it plays an essential role in light detection and cone phototransduction by rapidly decreasing intracellular levels of cGMP. There is a lot of evidence that in different species, PDE6C has a crucial role in the cone phototransduction cascade.
PDE6C related signaling pathway
Among its related pathways are Phosphodiesterases in neuronal function and Visual signal transduction: Cones.
PDE6C Related Diseases
Cone dystrophy type 4. An early-onset cone dystrophy. Cone dystrophies are retinal dystrophies characterized by progressive degeneration of the cone photoreceptors with preservation of rod function, as indicated by electroretinogram. The compound heterozygous in PDE6C mutations caused CD and the findings show that CD and ACHM constitute genetically and clinically overlapping phenotypes.
Achromatopsia 5. A form of achromatopsia, an ocular stationary disorder due to the absence of functioning cone photoreceptors in the retina. It is characterized by total colorblindness, low visual acuity, photophobia and nystagmus. ACHM5 inheritance is autosomal recessive. The homozygous or compound heterozygous mutations in PDE6C and PDE6H (OMIM #601190) lead to predominantly cone dysfunction disorders, such as achromatopsia.
Cone–rod dystrophy. An atypical phenotype reported in recent years with fewer PDE6C mutations is cone–rod dystrophy. PDE6C has been screened in cone–rod dystrophy cohorts and has been attributed to up to 1.8% of cases.
Biomedical Application of PDE6C Protein
Mechanism research model: An intron of the cone-specific Pde6c gene acted as an enhancer in human cones. This new transgenic human stem cell lines provide a tool for determining which cone developmental mechanisms are shared and distinct between mice and humans.
Case Study
Case Study 1: Tanja Grau, 2011
Mutations in the gene encoding the catalytic subunit of the cone photoreceptor phosphodiesterase (PDE6C) have been recently reported in patients with autosomal recessive inherited achromatopsia (ACHM) and early-onset cone photoreceptor dysfunction. Here the researchers present the results of a comprehensive study on PDE6C mutations including the mutation spectrum, its prevalence in a large cohort of ACHM/cone dysfunction patients, the clinical phenotype and the functional characterization of mutant PDE6C proteins. Twelve affected patients from seven independent families segregating PDE6C mutations were identified in the total patient cohort of 492 independent families. Eleven different PDE6C mutations were found including two nonsense mutations, three mutations affecting transcript splicing as shown by minigene assays, one 1 bp-insertion and five missense mutations. They also performed a detailed functional characterization of six missense mutations applying the baculovirus system to express recombinant mutant and wildtype chimeric PDE6C/PDE5 proteins in Sf9 insect cells. Purified proteins were analyzed using Western blotting, phosphodiesterase (PDE) activity measurements as well as inhibition assays by zaprinast and Pγ. Four of the six PDE6C missense mutations led to baseline PDE activities and most likely represent functional null alleles.
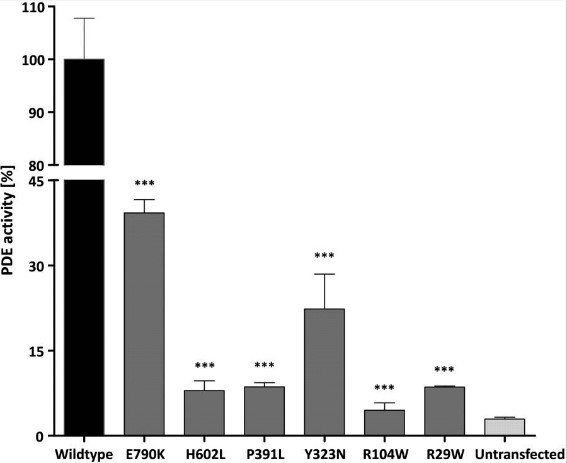
Fig1. Reduced or loss of catalytic activity of mutant PDE6C/PDE5 chimeras.
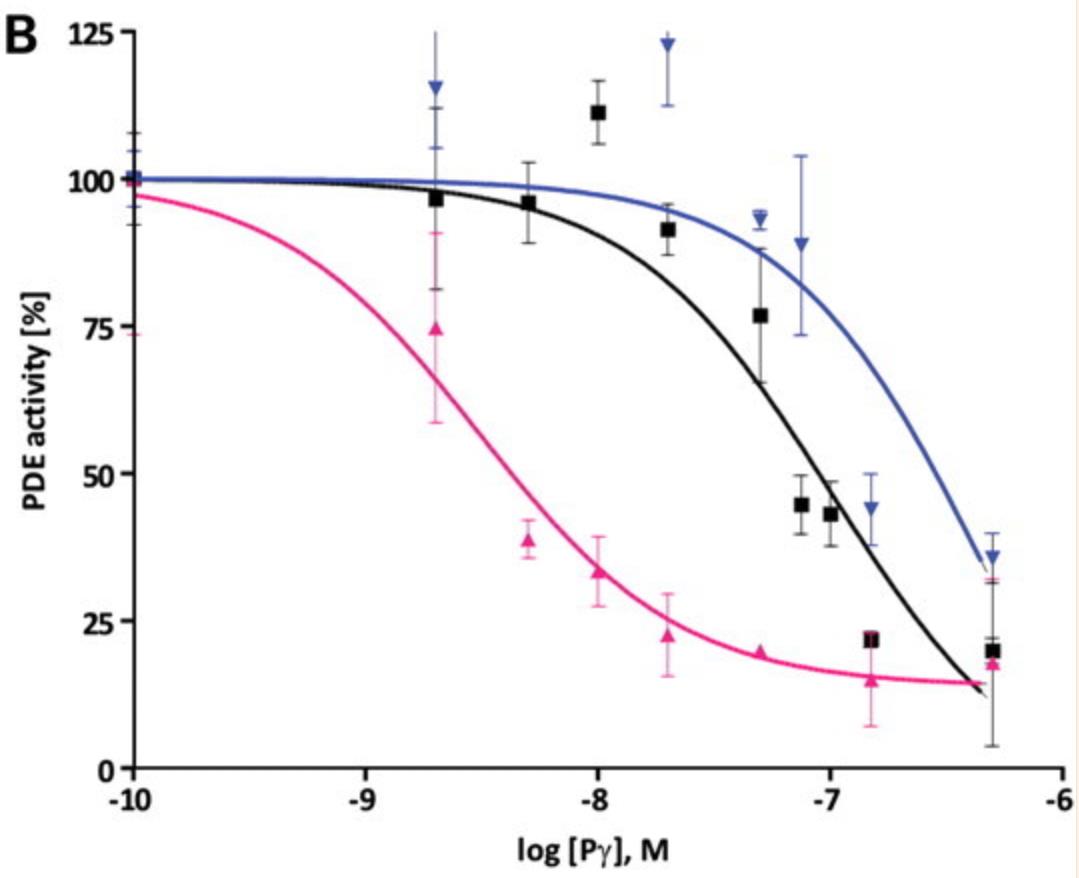
Case Study 2: Hakim Muradov, 2010
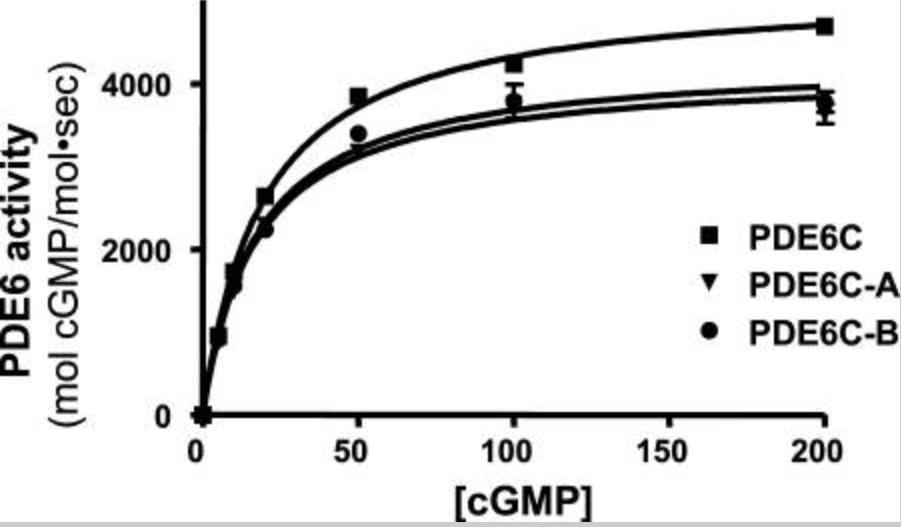
Phosphodiesterase-6 (PDE6) is the key effector enzyme of the phototransduction cascade in rods and cones. The catalytic core of rod PDE6 is a unique heterodimer of PDE6A and PDE6B catalytic subunits. The functional significance of rod PDE6 heterodimerization and conserved differences between PDE6AB and cone PDE6C and the individual properties of PDE6A and PDE6B are unknown. To address these outstanding questions, the researchers expressed chimeric homodimeric enzymes, enhanced GFP (EGFP)-PDE6C-A and EGFP-PDE6C-B, containing the PDE6A and PDE6B catalytic domains, respectively, in transgenic Xenopus laevis. Similar to EGFP-PDE6C, EGFP-PDE6C-A and EGFP-PDE6C-B were targeted to the rod outer segments and concentrated at the disc rims. PDE6C, PDE6C-A, and PDE6C-B were isolated following selective immunoprecipitation of the EGFP fusion proteins. All three enzymes, PDE6C, PDE6C-A, and PDE6C-B, hydrolyzed cGMP with similar K(m) (20-23 μM) and k(cat) (4200-5100 s(-1)) values. Likewise, the K(i) values for PDE6C, PDE6C-A, and PDE6C-B inhibition by the cone- and rod-specific PDE6 γ-subunits (Pγ) were comparable. Recombinant cone transducin-α (Gα(t2)) and native rod Gα(t1) fully and potently activated PDE6C, PDE6C-A, and PDE6C-B. In contrast, the half-maximal activation of bovine rod PDE6 required markedly higher concentrations of Gα(t2) or Gα(t1).
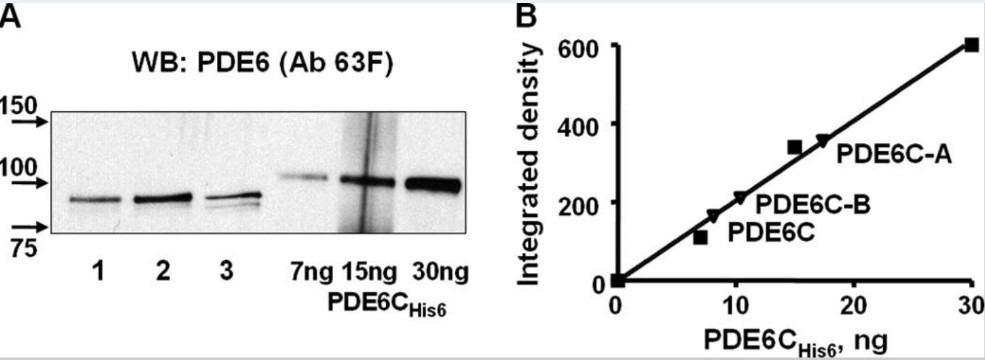
Fig3. Solubilization and quantification of PDE6C, PDE6C-A, and PDE6C-B (A and B are the catalytic core of rod heterodimer PDE6.).
Involved Pathway
PDE6C involved in several pathways and played different roles in them. We selected most pathways PDE6C participated on our site, such as Purine metabolism, which may be useful for your reference. Also, other proteins which involved in the same pathway with PDE6C were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Purine metabolism | AK5,TWISTNB,URAH,CECR1A,ENTPD1,PDE6A,ADSL,POLR2H,POLR1A,POLR3G |
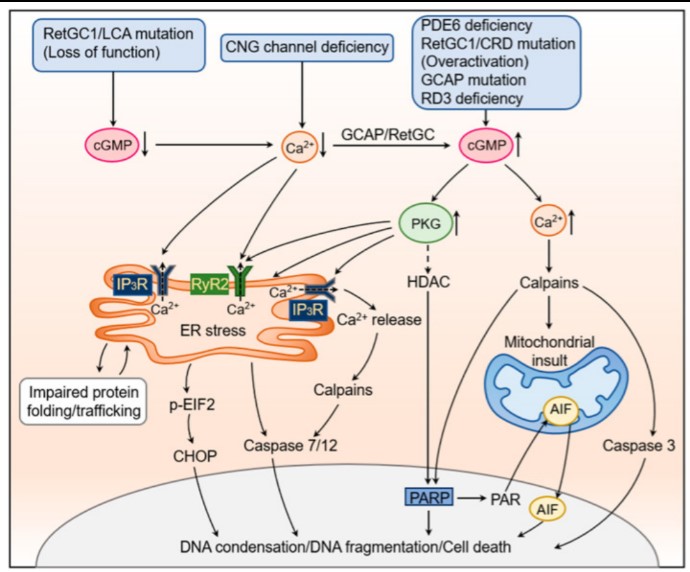
Fig1. The cellular and molecular mechanisms underlying cGMP signaling-induced photoreceptor degeneration.
Protein Function
PDE6C has several biochemical functions, for example, 3,5-cyclic-GMP phosphodiesterase activity,cGMP binding,metal ion binding. Some of the functions are cooperated with other proteins, some of the functions could acted by PDE6C itself. We selected most functions PDE6C had, and list some proteins which have the same functions with PDE6C. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| 3,5-cyclic-GMP phosphodiesterase activity | PDE6A,PDE10A,PDE6B,PDE11A,PDE6G,PDE9A,PDE6H,PDE5A |
| metal ion binding | ADAL,CYP2X9,EXTL2,TIMM10B,Car2,JAZF1B,NPEPPS,KALRN,ZNF668,ZFP513 |
| cGMP binding | PDE6G,PRKG2,CNGA5,PDE10A,CNGA4,CNGA2,PRKG1A,CNGA3,PDE6H,PDE11A |
Interacting Protein
PDE6C has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with PDE6C here. Most of them are supplied by our site. Hope this information will be useful for your research of PDE6C.
Resources
Related Services
Related Products
References



