Yeast Display Platform
Creative BioMart’s Yeast Display Platform leverages the power of Saccharomyces cerevisiae to facilitate directed evolution and protein engineering of antibodies, enzymes, and other functional proteins. By displaying target proteins on the yeast cell wall, our platform enables high-throughput selection of variants with enhanced binding, stability, or catalytic properties. Combining in vitro and in vivo mutagenesis with advanced screening technologies such as fluorescence-activated cell sorting (FACS), we provide a complete, one-stop solution from library construction to recombinant protein expression and purification. Our platform ensures precise, efficient, and scalable identification of high-performance protein variants for therapeutic, industrial, and research applications.
Understanding Yeast Display Technology
First introduced by Professor K. Dane Wittrup in 1997, yeast display has emerged as a robust protein engineering technique, particularly suitable for eukaryotic proteins. Saccharomyces cerevisiae serves as an ideal host due to its well-characterized cellular machinery, safety profile, and mannoprotein-rich cell wall, which facilitates surface display.
Proteins can be anchored to the yeast cell wall via N-terminal fusion (using Cwp1p, Cwp2p, Aga1p, Tip1p, Flo1p, Sed1p, YCR89w, Tir1) or C-terminal fusion (most commonly Aga2). The typical directed evolution workflow involves constructing a mutagenic library, transforming it into yeast, and screening for functional variants. Yeast also supports in vivo targeted mutagenesis and continuous evolution, enabling gene-specific diversification without compromising genomic integrity.
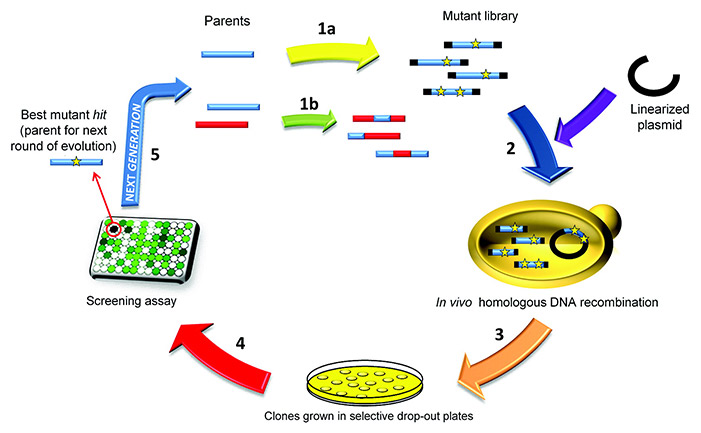
Figure 1. A typical directed evolution experiment using Saccharomyces cerevisiae as a eukaryotic host. (Gonzalez-Perez et al., 2012)
Applications of Yeast Display
|
Application Area |
Description |
Key Benefits |
|---|---|---|
|
Antibody Discovery & Engineering |
Isolation of high-affinity antibodies, affinity maturation, epitope mapping. |
Rapid identification of potent, specific antibodies for therapeutic or research use. |
|
Enzyme Engineering |
Directed evolution for improved activity, stability, substrate specificity. |
Optimized enzymes for industrial, clinical, or research applications. |
|
Ligand-Receptor Studies |
Screening peptides/proteins for receptor binding, mapping interaction sites. |
High-throughput functional analysis of protein–protein interactions. |
|
Therapeutic Protein Development |
Engineering proteins with enhanced stability, solubility, or reduced immunogenicity. |
Accelerates preclinical candidate optimization and functional validation. |
|
Synthetic Biology & Protein Design |
Generation of custom protein libraries, in vivo mutagenesis, continuous evolution. |
Exploration of novel binding motifs and enzymatic functions. |
|
Vaccine & Antigen Design |
Screening antigen variants for immunogenicity or epitope optimization. |
Supports design of epitope-focused vaccines and immunotherapies. |
What Our Yeast Display Platform Deliver
Creative BioMart provides a comprehensive yeast display service tailored to the discovery, optimization, and characterization of high-performance proteins:
- Design and engineering of protein or peptide display in S. cerevisiae.
- Construction of mutagenic libraries with genetic diversity generated via in vitro or in vivo methods.
- High-throughput screening, including FACS-based selection of functional variants.
- Recovery and characterization of selected clones.
- Recombinant expression and purification of evolved proteins for downstream applications.
- Flexibility to target binding affinity, catalytic activity, stability, or other desired functions.
Service Workflow

Service Features
|
Parameter |
Description |
|---|---|
|
Library Diversity |
Up to ~10⁷ unique variants per library. |
|
Mutagenesis Approaches |
Combination of in vitro mutagenesis, in vivo targeted mutagenesis, and in vivo continuous evolution (ICE). |
|
Display Strategies |
N-terminal and C-terminal fusions using multiple anchor proteins. |
|
Screening Methods |
High-throughput FACS, with optional complementary screening approaches. |
|
Protein Types |
Antibodies (VHH/nanobodies), enzymes, receptor fragments, and other functional peptides. |
|
Application Scope |
Therapeutics, enzyme optimization, biosensors, ligand-binding studies, and fundamental research. |
Why Choose Creative BioMart
- Validated Platform: Proven S. cerevisiae system optimized for protein surface display and library handling.
- High Library Diversity: Robust mutagenesis methods generate libraries with up to 10⁷ unique variants.
- Advanced Screening: FACS-enabled high-throughput selection ensures rapid identification of superior variants.
- Integrated Workflow: End-to-end service from library construction to recombinant protein production.
- Flexible Targeting: Supports a wide range of protein types, desired functions, and selection criteria.
- Experienced Team: Deep expertise in yeast genetics, protein engineering, and directed evolution technologies.
Case Studies and Real-World Applications
Case 1: Yeast display enables in vitro nanobody discovery
McMahon et al., 2018. doi:10.1038/s41594-018-0028-6
Camelid single-domain antibody fragments, or nanobodies, offer the specificity of conventional antibodies within a single 15-kDa VHH domain, making them valuable as biochemical tools and therapeutic agents. They are particularly useful in structural biology, stabilizing conformationally dynamic proteins such as GPCRs. Traditional nanobody discovery relies on animal immunization, limiting accessibility and throughput. To overcome this, a fully in vitro yeast surface display platform has been developed, enabling rapid identification of nanobodies, including conformationally selective binders to human GPCRs. The platform has been validated through antigen co-crystallization and is openly available for nonprofit research, expanding opportunities for structural and functional studies.
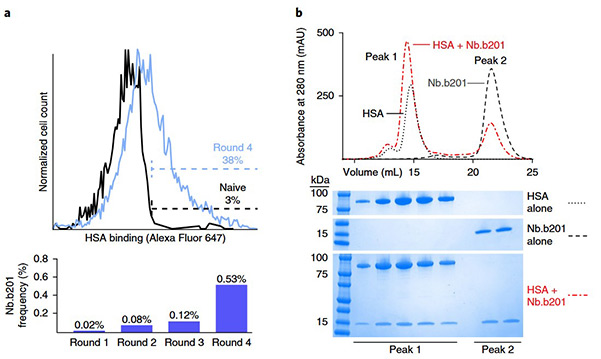
Figure 2. Validation of nanobody platform using HSA as the target antigen. a, Histogram of yeast library HSA binding, showing pre-selection (black) and after four rounds of selection (blue). The percentage of HSA-binding cells is indicated. The fraction of the total library composed by Nb.b201 was assessed by deep sequencing, showing progressive enrichment of this clone. b, Size-exclusion chromatography analysis confirmed the binding of purified recombinant Nb.b201 to HSA. (McMahon et al., 2018)
Case 2: Yeast display for enhanced bio-ethanol production
Yamakawa et al ., 2012. doi:10.1016/j.enzmictec.2012.03.005
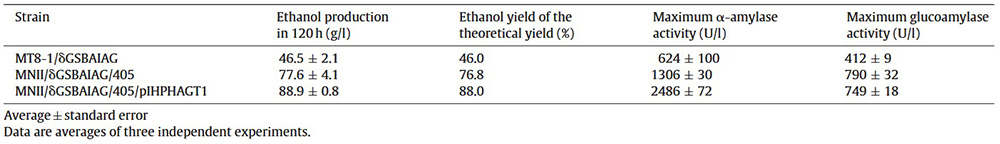
Using yeast surface display, a diploid Saccharomyces cerevisiae strain was engineered to co-display α-amylase and glucoamylase for efficient starch fermentation. The α-amylase construct was optimized for surface expression, producing 46.5 g/L ethanol from 200 g/L raw corn starch—1.5-fold higher than native α-amylase-displaying yeast. The co-displaying strain maintained enzyme activity over 23 repeated fermentation cycles, achieving an average ethanol productivity of 1.61 g/L/h and 76.6% of the theoretical yield. This approach allows easy reuse of yeast with surface-displayed amylases and demonstrates potential for cost-effective, industrial-scale bio-ethanol production from raw starch or whole grains.
Table 1. Ethanol production from 200 g/L of raw starch by glucoamylase and modified α-amylase co-displaying strains. (Yamakawa et al., 2012)
Customer Testimonials—Yeast Display Platform
“We collaborated with Creative BioMart to evolve a VHH nanobody library against a challenging tumor antigen. Their yeast display platform, combined with FACS-based high-throughput screening, enabled the rapid identification of high-affinity clones. The selected variants not only demonstrated superior binding but also retained stability under physiological conditions. Their expertise in library design and surface display significantly accelerated our preclinical program.”
— Director of Biologics R&D | Global Pharmaceutical Company
“Using Creative BioMart’s yeast display system, we engineered an industrial lipase for improved thermal stability. The team’s in vivo mutagenesis strategy generated a diverse library, and their high-throughput screening efficiently isolated top-performing variants. Recombinant protein characterization confirmed a 4-fold increase in activity at elevated temperatures, exceeding our expectations.”
— Senior Scientist | Industrial Enzyme Manufacturer
“Creative BioMart helped us identify high-specificity receptor-binding peptides through yeast surface display. Their integrated workflow—from library construction to clone recovery—streamlined our ligand discovery process. The selected candidates exhibited excellent selectivity and functional activity, providing a strong foundation for our biosensor development pipeline.”
— Principal Investigator | Biotech Start-up
“We leveraged Creative BioMart’s yeast display platform for affinity maturation of an antibody fragment. Their team designed an optimized library and applied iterative FACS selection, which yielded clones with over 10-fold improved affinity. The seamless integration with recombinant protein expression and purification saved us several months in development.”
— Head of Protein Engineering | Mid-sized Biopharma
FAQs About Our Yeast Display Service
-
Q: What types of proteins can be displayed on your yeast platform?
A: Our platform supports antibodies (including VHH/nanobodies), enzymes, receptor fragments, and other peptides, enabling directed evolution for binding, activity, or stability improvements. -
Q: What is the typical library diversity achievable?
A: We routinely generate libraries with up to ~10⁷ unique variants, using both in vitro and in vivo mutagenesis methods, including continuous evolution approaches. -
Q: How is high-throughput screening performed?
A: We use FACS-based selection to efficiently isolate top-performing clones, but complementary approaches can be applied depending on the target and desired function. -
Q: Can your system handle proteins that require eukaryotic folding or post-translational modifications?
A: Yes. Using S. cerevisiae as a eukaryotic host allows proper protein folding, disulfide bond formation, and other post-translational modifications, making it ideal for mammalian proteins. -
Q: How do you integrate yeast display with recombinant protein production?
A: Selected clones from our yeast display workflow can be transferred to recombinant expression systems for large-scale protein production, purification, and functional validation.
Other Resources
Related Services
- Protein Engineering Services
- Directed Evolution
- Protein Expression and Purification Services
- Yeast Expression Systems
- Mammalian Expression Systems
- Large-Scale Protein Production Service
- Epitope Mapping
- Protein Interaction Service
- Phage Display Platform
- Escherichia coli Display Platform
- Special Cell-based Display Platform
- Cell-free Display Platform
Related Products
References:
- Gonzalez-Perez D, Garcia-Ruiz E, Alcalde M. Saccharomyces cerevisiae in directed evolution: An efficient tool to improve enzymes. Bioengineered. 2012;3(3):174-179. doi:10.4161/bbug.19544
- McMahon C, Baier AS, Pascolutti R, et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol. 2018;25(3):289-296. doi:10.1038/s41594-018-0028-6
- Yamakawa S Ichi, Yamada R, Tanaka T, Ogino C, Kondo A. Repeated fermentation from raw starch using Saccharomyces cerevisiae displaying both glucoamylase and α-amylase. Enzyme and Microbial Technology. 2012;50(6-7):343-347. doi:10.1016/j.enzmictec.2012.03.005
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

