Histone Phosphorylation
Related Symbol Search List
- ATR
- AURKB
- BAZ1B
- CDK2
- Chk1
- CSNK2A1
- FYN
- GSG2
- JAK2
- ERK2
- MAPK14
- ERK1
- MAPK3
- MAPK8
- JNK2
- PAK2
- PKM2
- PRKAA1
- PRKCB
- PRKCD
- PRKDC
- RSK2
- MSK1
- MST1
- WEE1
Immunology Background
Background
Histone phosphorylation refers to the addition of phosphate groups to specific amino acid residues on histone proteins, which are structural components of chromatin. Histone phosphorylation is a reversible and dynamic post-translational modification that plays important roles in regulating chromatin structure, gene expression, and various cellular processes.
The function of histone phosphorylation can vary depending on the specific phosphorylated residue and the context in which it occurs. Here are some key functions of histone phosphorylation:
- Chromatin Remodeling: Histone phosphorylation can contribute to the relaxation or compaction of chromatin structure, influencing the accessibility of DNA to transcriptional machinery. Phosphorylation of histone H1 and H3 can lead to chromatin decondensation, facilitating gene expression.
- Transcriptional Regulation: Phosphorylation of specific histone residues can serve as a signal for transcriptional activation or repression. For example, phosphorylation of serine 10 on histone H3 (H3S10ph) is associated with gene activation, while phosphorylation of serine 28 on histone H3 (H3S28ph) is linked to gene repression.
- DNA Damage Response: Histone phosphorylation is a critical component of the cellular response to DNA damage. Phosphorylation of histone H2AX at serine 139 (γH2AX) is a well-known marker for DNA double-strand breaks and helps recruit DNA repair factors to the damaged sites.
- Mitosis and Chromosome Segregation: Histone phosphorylation is involved in regulating mitotic progression and proper chromosome segregation. Phosphorylation of histone H3 at serine 10 and serine 28 occurs during mitosis and is associated with chromosome condensation and sister chromatid resolution.
- Signaling Pathways: Histone phosphorylation can be regulated by various signaling pathways, including those activated by growth factors, hormones, and stress signals. It can serve as a mechanism for integrating extracellular signals with chromatin dynamics and gene expression.
- Epigenetic Memory: Histone phosphorylation can contribute to the establishment and maintenance of epigenetic memory. For instance, phosphorylation of histone H3 at serine 10 during cellular memory formation has been associated with the stabilization of gene expression patterns.
Overall, histone phosphorylation is a dynamic modification that contributes to the regulation of chromatin structure and gene expression, as well as various cellular processes. Its precise functions depend on the specific phosphorylated residue and the cellular context in which it occurs.
Histone Phosphorylation-Related Molecules
Several molecules are involved in the regulation and signaling of histone phosphorylation. These molecules play crucial roles in modifying histones and influencing gene expression. Here are some key molecules associated with histone phosphorylation:
Kinases
Kinases are enzymes that catalyze the addition of phosphate groups to histones. They are responsible for histone phosphorylation and can target specific residues on histone proteins. Examples of kinases involved in histone phosphorylation include:
- Aurora kinase: Phosphorylates histone H3 at serine 10 (H3S10) during mitosis and plays a role in chromosome condensation and segregation.
- Checkpoint kinases (CHK1 and CHK2): Phosphorylate histone H3 at serine 10 and serine 28 in response to DNA damage, contributing to DNA repair and cell cycle regulation.
- Protein kinase A (PKA): Phosphorylates histone H3 at serine 10 and serine 28, influencing gene expression and cellular processes.
Phosphatases
Phosphatases are enzymes that remove phosphate groups from histones, counterbalancing the action of kinases. They play a role in regulating the dynamics of histone phosphorylation. Examples of phosphatases involved in histone dephosphorylation include:
- Protein phosphatase 1 (PP1): Dephosphorylates histone H3 at serine 10 and serine 28, regulating gene expression and chromatin structure.
- Protein phosphatase 2A (PP2A): Involved in the dephosphorylation of histone H3 at serine 10 and serine 28, affecting gene expression and cellular processes.
Signaling Pathways
Various signaling pathways can regulate histone phosphorylation. These pathways transmit signals from extracellular stimuli to the nucleus, ultimately influencing the activity of kinases and phosphatases. Examples of signaling pathways involved in histone phosphorylation include:
- MAPK/ERK pathway: Activation of the MAPK pathway leads to the phosphorylation of histone H3 at serine 10, regulating gene expression and cell proliferation.
- PI3K/Akt pathway: The Phosphoinositide 3-kinase (PI3K) pathway and its downstream effector protein kinase B (Akt) can influence histone phosphorylation, affecting gene expression and cell survival.
Readers and Effectors
Proteins known as "readers" or "effectors" recognize and bind to specific histone phosphorylation marks, translating the histone code into functional outcomes. They can recruit other proteins or complexes to modulate chromatin structure and gene expression. Examples of readers and effectors of histone phosphorylation include:
- 14-3-3 proteins: These proteins bind to H3S10-phosphorylated histones and regulate gene expression, cell cycle progression, and apoptosis.
- BRD4: This bromodomain-containing protein recognizes acetylated histones and phosphorylated H3S10, playing a role in transcriptional activation and chromatin remodeling.
Representative Types and Members of Histone Phosphorylation
| Types | Details |
|---|---|
| Cyclin-Dependent Kinases (CDKs) |
|
| Aurora Kinase |
|
| Mitogen-Activated Protein Kinase (MAPK ) |
|
| DNA Damage Responsive Kinase |
|
| Protein Kinase C (PKC) |
|
| Other Kinases |
|
These molecules collectively contribute to the dynamic regulation of histone phosphorylation, influencing chromatin structure, gene expression, and various cellular processes. Understanding their interactions and functions is crucial for unraveling the complex mechanisms underlying histone phosphorylation.
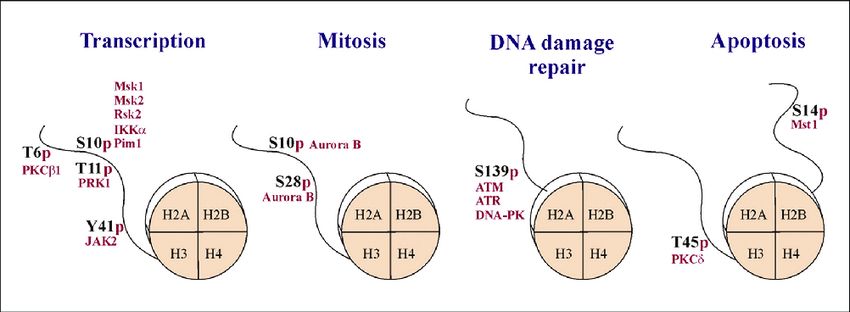 Fig.1 Histone phosphorylation is implicated in multiple cellular processes. (Cohen I, et al., 2011)
Fig.1 Histone phosphorylation is implicated in multiple cellular processes. (Cohen I, et al., 2011)Histone Phosphorylation and Disease Association
Histone phosphorylation plays a significant role in the occurrence and progression of various diseases. Let's explore the association between histone phosphorylation and diseases such as cancer, neurological disorders, and cardiovascular diseases, along with the related research advancements.
Cancer
Aberrant histone phosphorylation is closely associated with the development of cancer. Phosphorylation modifications can alter chromatin structure and function, thereby affecting gene transcription and expression. Here are some research advancements related to histone phosphorylation in cancer:
- Abnormal expression of H3K27 phosphorylation is linked to dysregulation of the transcriptional repressor Polycomb group protein complex (PRC2) in various cancers, leading to aberrant gene activation or repression, promoting cancer cell proliferation and metastasis.
- H3S10 phosphorylation has been found to be upregulated in multiple cancers, including breast cancer, lung cancer, and colorectal cancer. This upregulation is associated with cancer cell proliferation, invasion, and metastasis.
- H2AX serves as a marker protein in DNA damage response, and its phosphorylation form (γH2AX) is activated during DNA double-strand breaks. Dysregulation of DNA damage repair mechanisms in cancer cells leads to abnormal expression of γH2AX, which is associated with cancer occurrence and treatment response.
Neurological Disorders
Abnormal histone phosphorylation is also associated with the occurrence and progression of neurological disorders. Research advancements related to histone phosphorylation in neurological disorders include:
- H3S10 phosphorylation plays a critical role in neuronal development and synaptic plasticity, and other neurological functions. Abnormal H3S10 phosphorylation is associated with neurological disorders such as Parkinson's disease, Alzheimer's disease, and schizophrenia.
- H3T11 phosphorylation is a neuroactivity-dependent modification involved in memory formation and synaptic plasticity. Studies have found that abnormal expression of H3T11 phosphorylation is associated with neurological disorders such as Alzheimer's disease and hippocampal dysfunction.
Cardiovascular Diseases
Histone phosphorylation also plays a role in cardiovascular diseases. It contributes to the regulation of gene expression in cardiac cells, influencing processes such as cardiac hypertrophy, remodeling, and arrhythmias. Abnormal histone phosphorylation patterns have been observed in conditions such as heart failure, atherosclerosis, and cardiac arrhythmias. Here are some research advancements related to histone phosphorylation in cardiovascular diseases:
- H3S10 phosphorylation has been found to be aberrantly upregulated in cardiac cell proliferation and cardiac hypertrophy, and other cardiovascular diseases. Studies suggest that regulation of H3S10 phosphorylation contributes to the pathogenesis of these conditions.
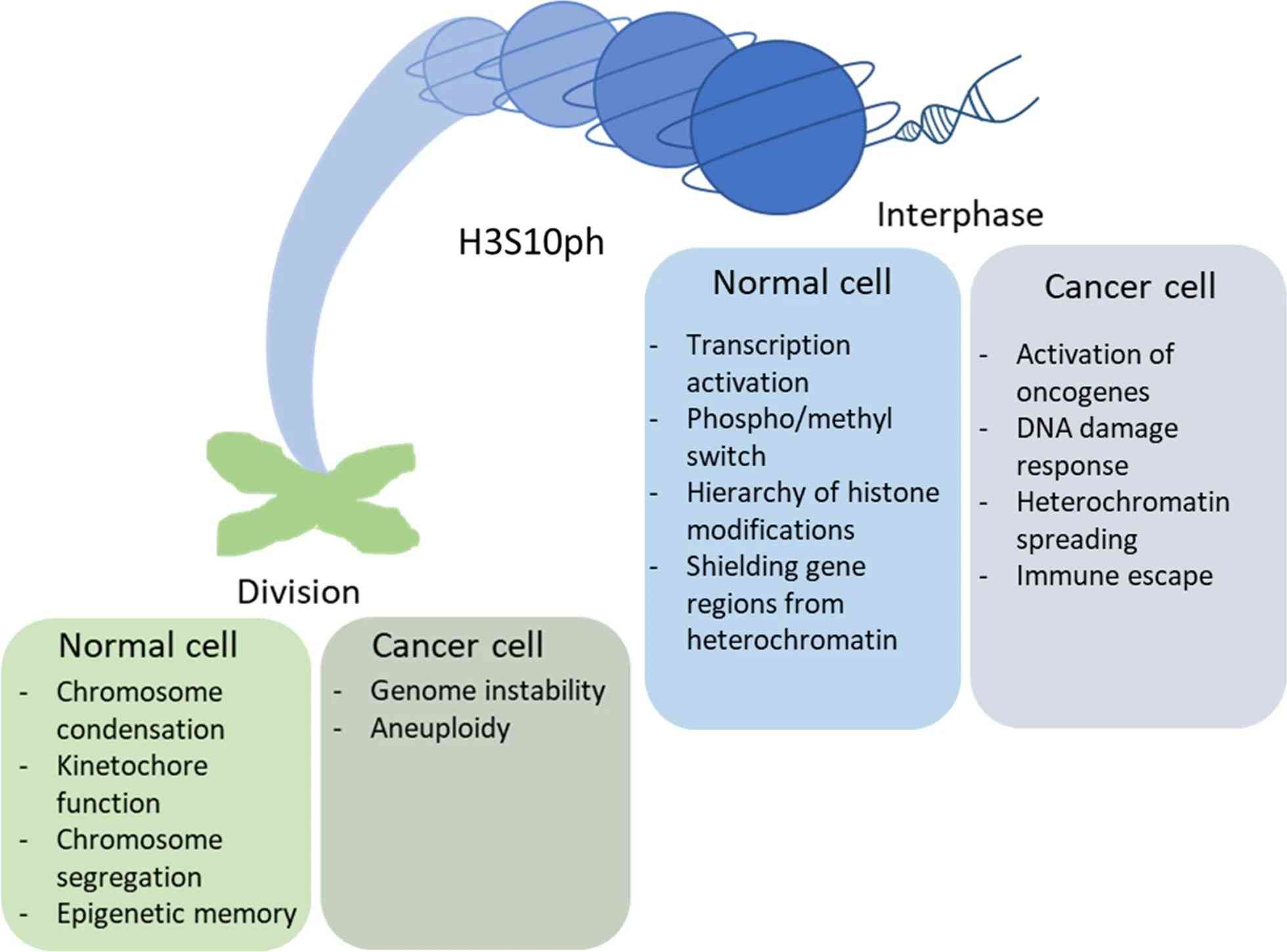 Fig.2 Effect of H3S10 phosphorylation on chromatin activity in normal and cancer cells. (Komar D, et al., 2020)
Fig.2 Effect of H3S10 phosphorylation on chromatin activity in normal and cancer cells. (Komar D, et al., 2020)Case Study
Case 1: Moura DS, Campillo-Marcos I, Vázquez-Cedeira M, Lazo PA. VRK1 and AURKB form a complex that cross inhibit their kinase activity and the phosphorylation of histone H3 in the progression of mitosis. Cell Mol Life Sci. 2018;75(14):2591-2611.
During the cell cycle, VRK1 and AURKB play important roles in histone phosphorylation and chromatin condensation. VRK1 is expressed throughout the cell cycle but reaches its peak activity in the G2/M phase, where it is required for chromosome condensation. On the other hand, AURKB is specifically expressed during mitosis, mainly in metaphase and anaphase.
The formation of the VRK1-AURKB complex is mainly observed after nocodazole release and coincides with the loss of H3 phosphorylations and phosphorylated Rb, which may signal the completion and exit of mitosis. The immunoprecipitation analysis confirms the timing of the AURKB-VRK1 complex formation and the loss of H3T3ph followed by H3S10ph.
Immunofluorescence studies further support these findings. H3-Thr3 phosphorylation is localized in the periphery during prophase and moves to condensed chromosomes in prometaphase. However, it is lost in anaphase when the VRK1-AURKB interaction can occur. In contrast, H3-Ser10 phosphorylation remains associated with chromatin throughout mitosis.
The timing of these phosphorylation patterns observed in the cell cycle phases aligns with the patterns detected in the population through immunoblots. VRK1 is not detected on chromatin in the later phases of the cell cycle, as it is ejected following the phosphorylation of H3-Thr3 during chromatin condensation in mitosis.
In summary, VRK1 and AURKB, through their interaction and phosphorylation of specific histone H3 residues, contribute to the regulation of chromatin condensation and progression through the cell cycle.
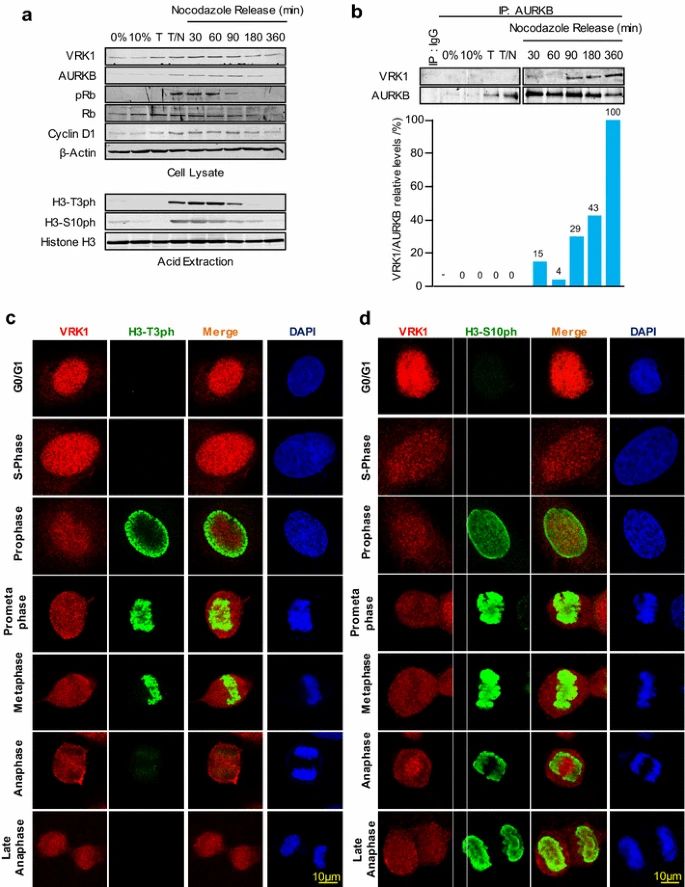 Fig.1 VRK1-AURK complex and histone H3 phosphorylation in cell cycle progression.
Fig.1 VRK1-AURK complex and histone H3 phosphorylation in cell cycle progression.Case 2: Lashen A, Algethami M, Alqahtani S, Shoqafi A, Sheha A, Jeyapalan JN, Mongan NP, Rakha EA, Madhusudan S. The clinicopathological significance of the cyclin D1/E1–cyclin-dependent kinase (CDK2/4/6)–retinoblastoma (RB1/pRB1) pathway in epithelial ovarian cancers. International Journal of Molecular Sciences. 2024; 25(7):4060.
The study focuses on evaluating the prognostic and predictive significance of the CDK2/4/6–cyclin D1/E1–pRB1 axis in clinical ovarian cancers. The expression levels of CDK2, CDK4, and CDK6 were investigated in 300 ovarian cancer samples and correlated with clinicopathological parameters and patient outcomes.
The immunohistochemical staining revealed both nuclear and cytoplasmic expression of CDK2. High nuclear CDK2 expression was observed in 28% of ovarian cancer cases, while high cytoplasmic CDK2 expression was observed in the same percentage of cases. CDK4 expression was analyzed using X-tile software, and it was dichotomized into high and low expression groups. High nuclear CDK4 expression was observed in 16% of tumors, while high cytoplasmic CDK4 expression was observed in 31% of tumors. Both nuclear and cytoplasmic staining for CDK4 was observed in the samples. For CDK6, low nuclear expression was found in 74% of patients, while high nuclear expression was observed in 26% of patients. In terms of cytoplasmic expression, 91% of patients had low CDK6 expression, while 9% had high CDK6 expression.
These findings provide insights into the expression patterns of CDK2, CDK4, and CDK6 in ovarian cancers and their potential significance as prognostic and predictive markers.
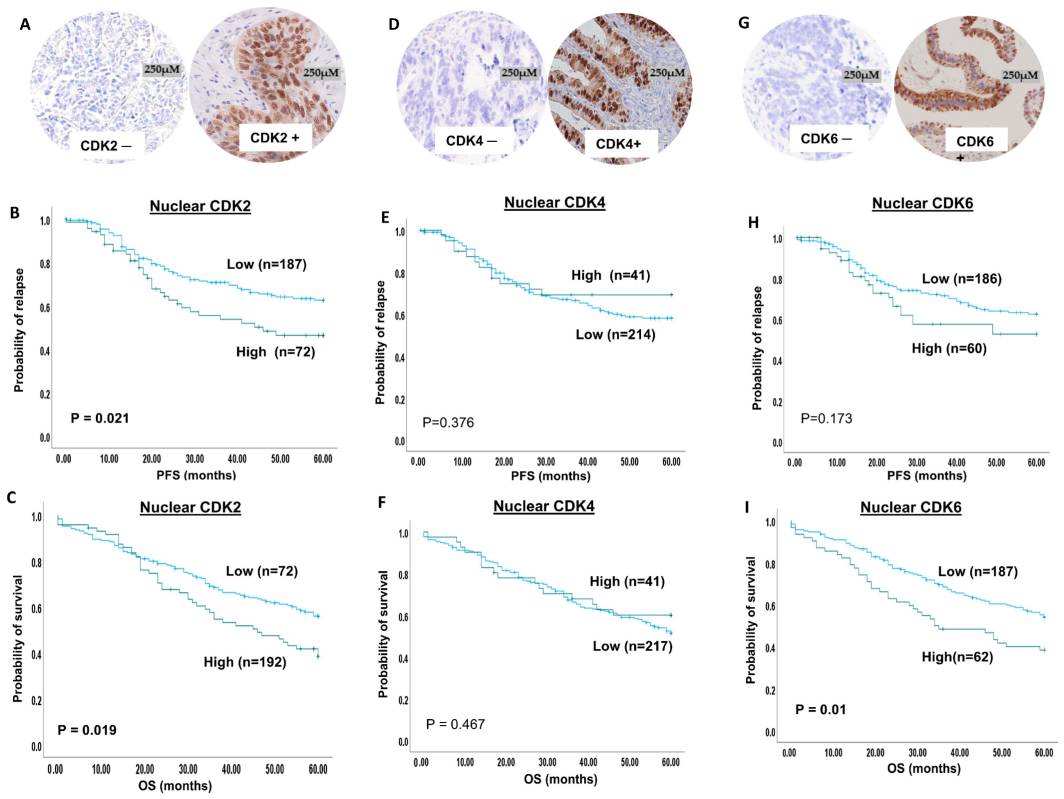 Fig.2 CDK2, 4 and 6 immunohistochemical expression in ovarian cancers.
Fig.2 CDK2, 4 and 6 immunohistochemical expression in ovarian cancers.Related References
- Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7(10):1098-1108.
- Komar D, Juszczynski P. Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin Epigenetics. 2020;12(1):147.
- Cohen I, Poręba E, Kamieniarz K, Schneider R. Histone modifiers in cancer: friends or foes?. Genes Cancer. 2011;2(6):631-647.

