H3 Family
Related Symbol Search List
- HIST1H3A
- HIST1H3B
- HIST1H3C
- HIST1H3D
- HIST1H3E
- HIST1H3F
- HIST1H3G
- HIST1H3H
- HIST1H3I
- HIST1H3J
- HIST2H3C
- HIST3H3
Immunology Background
Background
The histone H3 family is a group of proteins that are part of the histone protein family. Histones play a central role in packaging DNA into a compact and organized structure called chromatin, which is essential for regulating gene expression and maintaining genome stability. Histone H3 is one of the core histones and is highly conserved across species. Here is an introduction to the concept, structure, and function of the histone H3 family:
The histone H3 family refers to a group of closely related proteins that share structural and functional similarities. These proteins are encoded by different genes and exhibit sequence variations, post-translational modifications, and expression patterns that contribute to their specific functions within the context of chromatin.
Structure
Histone H3 proteins have a conserved structure consisting of a globular domain and an N-terminal tail. The globular domain forms part of the histone octamer core, which together with the histone H4, H2A, and H2B proteins, makes up the core nucleosome structure. The N-terminal tail extends outward from the nucleosome and is subjected to various post-translational modifications, such as acetylation, methylation, phosphorylation, and ubiquitination. These modifications can influence chromatin structure and gene expression.
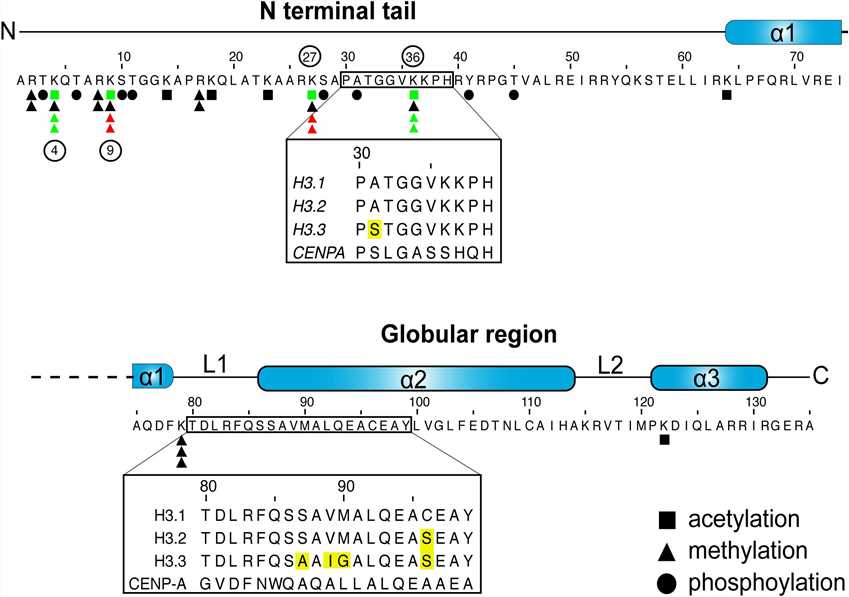 Fig.1 H3 sequence and posttranslational modifications. (Scott WA, et al., 2020)
Fig.1 H3 sequence and posttranslational modifications. (Scott WA, et al., 2020)Functions
The histone H3 family plays critical roles in chromatin organization, gene regulation, and other cellular processes. Some key functions include:
| Functions | Details |
|---|---|
| Chromatin compaction | Histone H3 proteins, as part of the nucleosome core, help package DNA into a highly compact and organized structure, facilitating the efficient storage and transmission of genetic information. |
| Gene regulation | Modifications of the histone H3 N-terminal tail, such as acetylation, methylation, and phosphorylation, act as important epigenetic marks that regulate gene expression. These modifications recruit various effector proteins that influence chromatin accessibility and the binding of transcription factors, ultimately controlling gene activation or repression. |
| Chromatin remodeling | Histone H3 variants, such as H3.3 and H3.1, are involved in dynamic chromatin remodeling processes. These variants are incorporated into specific genomic regions during DNA replication, DNA repair, and transcriptional activation, contributing to the dynamic regulation of chromatin structure and gene expression. |
| Epigenetic inheritance | Histone H3 modifications can be inherited across cell divisions and even between generations, contributing to the maintenance of gene expression patterns and cellular identity. |
| Genome stability | Histone H3 proteins play a role in maintaining genome integrity by promoting proper DNA replication, DNA repair, and chromosome segregation. |
The histone H3 family's diverse functions and regulatory roles in chromatin make it a subject of intensive research. Understanding the specific mechanisms by which histone H3 proteins and their modifications regulate gene expression and cellular processes is crucial for unraveling the complexities of epigenetic regulation and its impact on development, disease, and cellular homeostasis.
Members and Variants of the H3 Family
There are several members and variants of the histone H3 family, which are distinguished by post-translational modifications and sequence variations. Here are some of the well-known members and variants:
| Members and variants | Details |
|---|---|
| Histone H3.1 | Histone H3.1 is a canonical histone variant expressed during the S phase of the cell cycle. It is highly conserved and is incorporated into chromatin during DNA replication. |
| Histone H3.2 | Histone H3.2 is another canonical histone variant that is expressed throughout the cell cycle. It shares high sequence similarity with H3.1 and is also involved in the formation of nucleosomes. |
| Histone H3.3 | Histone H3.3 is a replication-independent histone variant that is expressed throughout the cell cycle. Unlike H3.1 and H3.2, H3.3 is incorporated into chromatin outside of DNA replication and is involved in various processes such as gene expression, DNA repair, and embryonic development. |
| Histone H3.4 | Histone H3.4 is a replication-independent histone variant with a similar function to H3.3. It is expressed in a tissue-specific manner and is involved in gene regulation and chromatin dynamics. |
| Histone H3.5 | Histone H3.5 is a less-studied variant that is expressed in specific cell types, such as embryonic stem cells. Its precise role and functions are still being investigated. |
| CenH3 (Centromeric Histone H3) | CenH3 is a specialized histone variant that is found specifically at centromeres, the specialized regions of chromosomes that are important for chromosome segregation during cell division. CenH3 replaces canonical H3 at centromeres and is crucial for proper kinetochore assembly. |
| Histone H3K4me3 (Histone H3 Lysine 4 Trimethylation) | This is a post-translational modification of histone H3 where a methyl group is added to the lysine 4 residue. H3K4me3 is associated with active gene transcription and is often found at the promoters of actively transcribed genes. |
| Histone H3K9me3 (Histone H3 Lysine 9 Trimethylation) | This is another post-translational modification of histone H3 where three methyl groups are added to the lysine 9 residue. H3K9me3 is associated with gene silencing and heterochromatin formation. |
| H3K27me3 (Histone H3 Lysine 27 Trimethylation) | This post-translational modification involves the addition of three methyl groups to the lysine 27 residue of histone H3. H3K27me3 is associated with gene repression and plays a role in cellular differentiation and development. |
| H3T (Histone H3 Threonine Phosphorylation) | This is a post-translational modification of histone H3 where a phosphate group is added to the threonine residue. H3T phosphorylation is associated with various cellular processes, including DNA damage response and cell cycle regulation. |
| H3S (Histone H3 Serine Phosphorylation) | H3S phosphorylation is another post-translational modification of histone H3, where a phosphate group is added to the serine residue. It is involved in signaling pathways that regulate gene expression and chromatin remodeling. |
The histone H3 family also includes several variants, such as variants HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J in the HIST1 cluster, variant HIST3H3 in the HIST3 cluster, and variant HIST2H3C in the HIST2 cluster. These variants are involved in nucleosome formation, chromatin organization, gene regulation, and DNA packaging. They play a vital role in maintaining chromatin structure, regulating gene expression, and ensuring genome stability.
These are some of the well-known members and variants of the histone H3 family, as well as specific post-translational modifications. Each member and modification contributes to the regulation of chromatin structure and gene expression and plays a key role in various cellular processes.
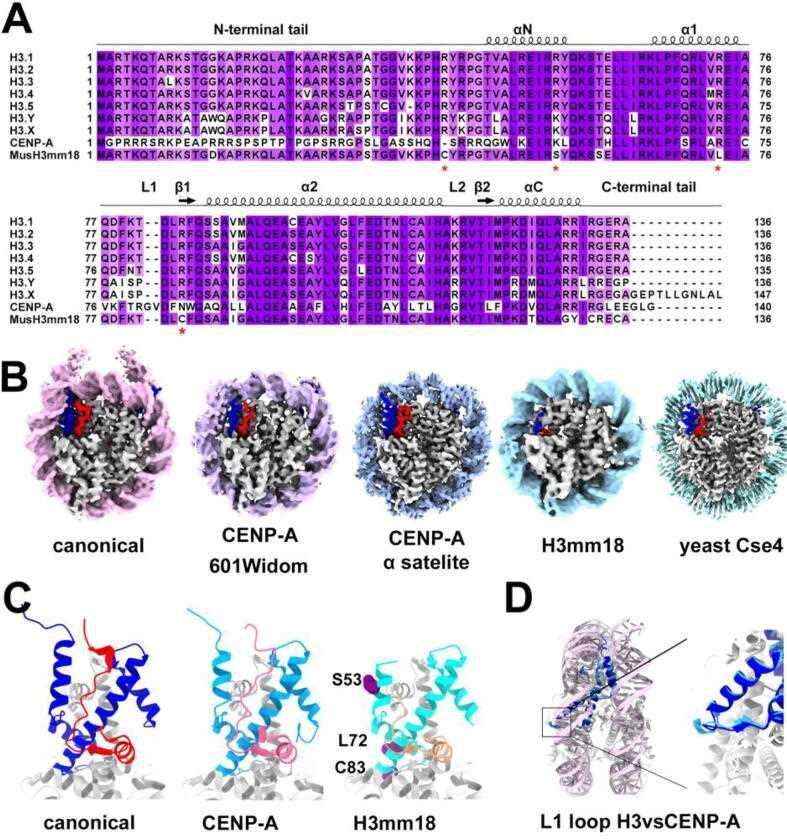 Fig.2 Structural diversity of histone H3 variants, CENP-A and H3mm18. (Sokolova V, et al., 2022)
Fig.2 Structural diversity of histone H3 variants, CENP-A and H3mm18. (Sokolova V, et al., 2022)Role of Histone H3 Family in Disease Development
Cancer
Histone H3 mutations have been identified in various types of cancers. For instance, mutations in the genes encoding histone H3.3 (H3F3A) and H3.1 (HIST1H3B) have been found in pediatric gliomas, including diffuse intrinsic pontine glioma (DIPG) and midline gliomas. These mutations affect specific amino acids in the histone H3 tail, leading to altered chromatin structure and gene expression patterns. Histone H3 mutations have also been identified in other cancers, such as chondroblastoma and giant cell tumors.
Epigenetic Modulators
The histone H3 family and its modifications are targets for epigenetic modulators that have shown promise in cancer therapy. Small molecules targeting enzymes involved in histone modifications, such as histone methyltransferases and histone deacetylases, have been developed as potential anti-cancer drugs. These inhibitors can modulate histone H3 modifications and chromatin structure, leading to changes in gene expression and potentially affecting tumor growth and survival.
Epigenetic Dysregulation in Neurological Disorders
Epigenetic alterations, including changes in histone H3 modifications, have been implicated in various neurological disorders. For example, in Alzheimer's disease, aberrant histone acetylation and methylation patterns have been observed, affecting the expression of genes involved in synaptic plasticity and memory formation. Similarly, histone H3 modifications have been linked to neurodegenerative diseases such as Parkinson's disease and Huntington's disease, contributing to disease progression.
Immunological Disorders
Histone H3 modifications play a role in immune cell development, differentiation, and immune response regulation. Dysregulation of histone H3 modifications has been associated with autoimmune diseases, such as systemic lupus erythematosus (SLE) and rheumatoid arthritis. Altered histone H3 modifications can affect the expression of immune-related genes, leading to aberrant immune responses and inflammation.
Therapeutic Targeting of Histone H3
The identification of histone H3 mutations and dysregulated histone H3 modifications in diseases has opened up opportunities for developing targeted therapies. Researchers are exploring the use of small molecules, such as inhibitors of histone-modifying enzymes, to restore normal histone H3 modifications and gene expression patterns. Additionally, strategies targeting specific histone H3 mutations, such as inhibitors that selectively target mutant histone H3 proteins, are being investigated as potential therapeutic approaches.
It's important to note that the field of epigenetics and histone research is rapidly evolving, and new findings continue to emerge. Ongoing research aims to deepen our understanding of the roles of the histone H3 family in disease development and identify novel therapeutic strategies based on this knowledge.
Case Study
Case 1: Gleason RJ, Guo Y, Semancik CS, Ow C, Lakshminarayanan G, Chen X. Developmentally programmed histone H3 expression regulates cellular plasticity at the parental-to-early embryo transition. Sci Adv. 2023;9(14):eadh0411.
In C. elegans, zygotic transcription begins at the 4-cell stage, while gastrulation starts at the 26-cell stage. In early embryogenesis, there is low expression of class I histone H3 genes (his-45, his-55, his-59, his-63) and high expression of the class II histone H3 gene his-6. However, upon gastrulation initiation, there is a significant increase in the expression of all 15 histone H3 genes. The expression timing of the H3 genes within the same class is coordinated, suggesting coexpression regulation. The highly expressed class II H3 genes are located on chromosome Ch V, while the germline-expressed class I H3 genes are on a different chromosome Ch IV. Notably, the genes encoding H3.3 (his-71 and his-72) do not show significant expression changes during gastrulation. The transition from an H3.3-enriched epigenetic landscape in late germline and early embryos to an H3-enriched epigenome in late embryos is specific to somatic lineage cells, while the P lineage for primordial germ cells maintains H3.3 enrichment throughout embryogenesis.
In summary, the study observed differential expression patterns of histone H3 genes during C. elegans embryogenesis, with distinct expression profiles for class I and class II H3 genes. The findings suggest developmental coordination and potential cis-regulatory mechanisms for the coexpression patterns of different classes of H3 genes. Additionally, there is a global change in the epigenome from H3.3-enriched to H3-enriched in somatic lineage cells during late embryogenesis.
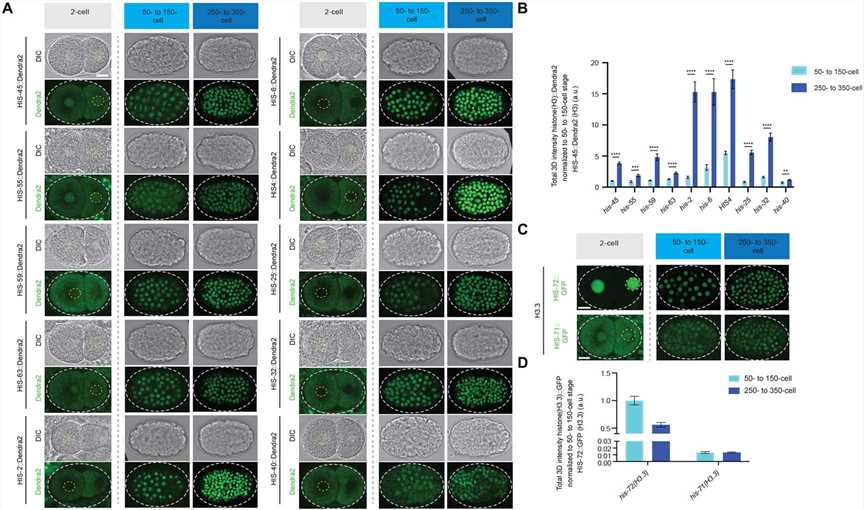 Fig.1 Expression patterns of all histone H3 gene clusters and H3.3 genes during early embryonic development.
Fig.1 Expression patterns of all histone H3 gene clusters and H3.3 genes during early embryonic development.Case 2: Kawamura M, Funaya S, Sugie K, Suzuki MG, Aoki F. Asymmetrical deposition and modification of histone H3 variants are essential for zygote development. Life Sci Alliance. 2021;4(8):e202101102.
The study investigated the distribution of the histone modification H3K27me3 in H3 overexpression variants. It was found that H3K27me3 signal was present throughout the maternal pronucleus, with higher overall levels than in the paternal pronucleus. However, H3K27me3 was specifically detected in the perinucleolar region of paternal pronuclei. Surprisingly, no reduction in H3K27me3 levels was observed in the perinucleolar region of paternal pronuclei in H3.1 and H3.2 overexpression embryos, despite a decrease in H3.3 levels in the paternal pronucleus. The findings suggested that the introduced H3.1 and H3.2 acquired the K27me3 modification in the paternal pronuclei.
The study hypothesized that the ectopic methylation of H3.1 and H3.2 at the K27me3 residue in the paternal perinucleolar region of H3.1/2 overexpression embryos could cause a delay in DNA replication. This hypothesis was based on previous studies that demonstrated the impact of histone modifications by microinjecting embryos with cRNA encoding H3 variants with specific amino acid substitutions. The findings suggested that the methylation of K27 in H3.1 and H3.2 within the paternal perinucleolar chromatin could lead to developmental failure.
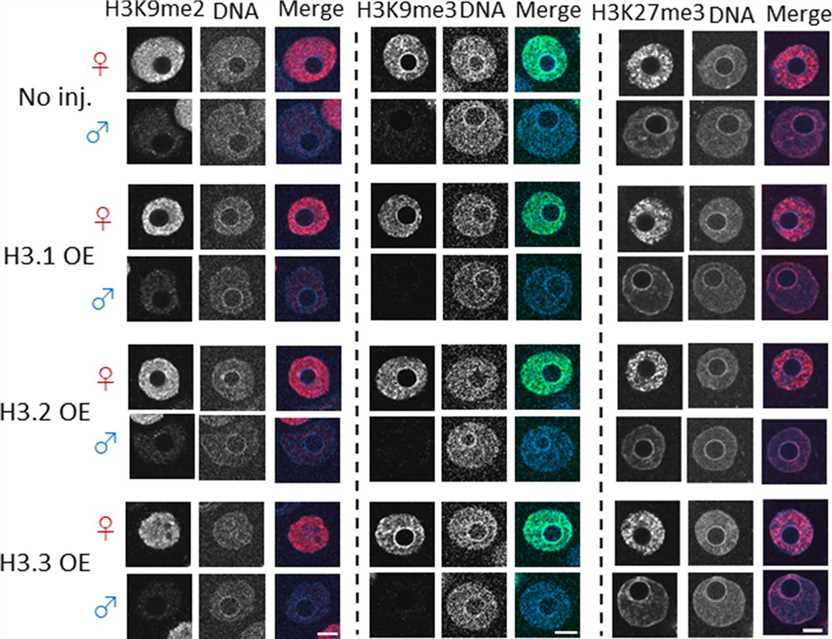 Fig.2 Confocal images showing the effect of forced nuclear incorporation of H3.1, H3.2, and H3.3 on H3K9me2/3 and H3K27me3.
Fig.2 Confocal images showing the effect of forced nuclear incorporation of H3.1, H3.2, and H3.3 on H3K9me2/3 and H3K27me3.References
- Ray-Gallet D, Almouzni G. The histone H3 family and its deposition pathways. Adv Exp Med Biol. 2021;1283:17-42.
- Scott WA, Campos EI. Interactions with histone H3 & tools to study them. Front Cell Dev Biol. 2020;8:701.
- Sokolova V, Sarkar S, Tan D. Histone variants and chromatin structure, update of advances. Comput Struct Biotechnol J. 2022;21:299-311.

