Guide for Mass Spectrometry Identification of Samples Separated by Liquid Chromatography
As one of the most powerful separation and analysis technologies in the field of chemistry, liquid chromatography (LC) has developed rapidly since the 1970s. Both in terms of fundamental chromatography theory and instrument performance, it has been greatly improved and improved. LC exhibits high separation efficiency and accurate quantification in the analysis of complex samples. With the continuous deepening of life science research, LC has become an extremely important and essential means of separation and analysis. However, the qualitative analysis ability of LC is relatively weak, usually only based on the retention characteristics of each component, which poses certain difficulties in complex sample analysis, especially in the qualitative analysis of unknown components. The liquid chromatography-mass spectrometry technology organically combines the high separation efficiency of liquid chromatography with the high sensitivity and strong structural analysis ability of mass spectrometry. After the sample is separated by liquid chromatography, it is identified using the results of mass spectrometry analysis, making it possible to separate and detect complex samples.
This section takes the method of combining nanoliter (nL) level liquid chromatography (Nano LC) with mass spectrometry as an example to explain the method of separating peptide targets using a liquid chromatography column after pancreatic enzyme hydrolysis, and then detecting them through MALDI-TOF-TOF mass spectrometry. The obtained data is analyzed using ProteinPilotTM software to identify the components of egg white matter.
Liquid-liquid partition chromatography is a separation method based on the difference in relative solubility of the measured substance between the stationary and mobile phases, by partitioning solutes between the two phases. According to the polarity difference between the stationary phase and the mobile phase, it can be divided into normal phase chromatography and reverse phase chromatography. The former uses silica gel or polar bonded phase as the stationary phase, and non-polar solvent as the mobile phase. The latter is an alkyl-bonded phase based on silica gel as the stationary phase, and a polar solvent as the mobile phase, suitable for the separation of non-polar compounds. Reverse liquid chromatography is commonly used for the separation of biological macromolecules such as proteins, peptides, and nucleic acids. Its separation mechanism, due to the hydrophobicity of the solid-phase carrier, can undergo strong or weak interactions based on the hydrophobicity of the separated substance molecules in the mobile phase, thus causing different molecules to separate from each other in the reverse phase column. The interaction between sample molecules with weaker hydrophobicity and the stationary phase is weaker, resulting in faster outflow; On the contrary, molecules with relatively strong hydrophobicity have strong interactions with the stationary phase, resulting in a relatively long retention time in the column.
After the protein sample is hydrolyzed by trypsin, a nano-upgraded HC reverse chromatography column (packed with octadecylsilane) is used. The chromatography column is a stationary phase, and the mobile phase is acetonitrile and water. By adjusting the ratio of acetonitrile to water, the polarity of the mobile phase is changed, thereby changing the distribution coefficient of peptide segments between the stationary phase and the mobile phase, and eluting the corresponding peptide segments. At the same time, mass spectrometry analysis was performed on the target peptide segments of graded elution to obtain target protein information.
1. Main Instruments and Equipment
Centrifuge, liquid chromatograph, mass spectrometer, sample target.
2. Material
A collection of total proteins or multiple proteins.
3. Main reagents*1
| (1) Chanel I | |
| Mobile phase A | 2% acetonitrile, 0.1% TFA. |
| Mobile phase B | 98% acetonitrile, 0.1% TFA. |
| (2) Substrate: 5mg/mL CHCA | |
| 5mg/mL | CHCA |
| 70% | Acetonitrile |
| 0.1% | TFA |
| (3) Chanel II 2% acetonitrile | |
1. Sample purification. Wash the protein with acetone pre-cooled at -20°C to remove impurities.
2. Trypsin digestion of the sample*2
3. Liquid chromatography column flushing and equilibration
Rinse*3: Rinse with 100% mobile phase B at a flow rate of 2μL/min for 24hrs.
Equilibration*4:
ChanelⅠ: Equilibrated with 95% mobile phase A and 5% mobile phase B at a flow rate of 2μL/min for 20min.
ChanelⅡ: Equilibrated with 100% mobile phase A, flow rate 2μL/min for 20min.
4. Liquid chromatography parameter setting:
According to the peak time*5 of the sample, set the corresponding parameters.
Select the column: Chromolith CapRod Cl18 column, 150 mm×0.1mm.
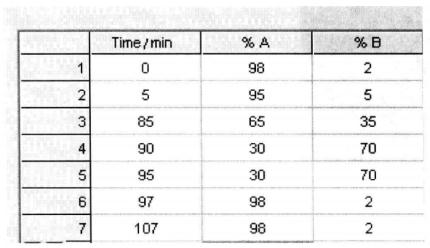
Setting of mobile phase and sample collection time*6:
| 85 min | 5% mobile phase ~ 35% mobile phase B |
| 5 min | 35% mobile phase ~ 70% mobile phase B |
| Flow rate | 2μL/min |
| Substrate flow rate | 2μL/min |
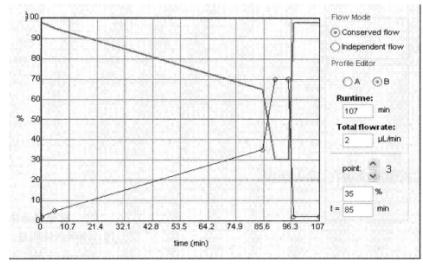
5. Point sample parameter setting*7: Point target speed is set to 0.7s/sample point.

6. Sample loading. Put the digested protein sample into the inner tube of the sample loading bottle.
7. Run the instrument according to the above settings to complete the sample spotting:
8. Mass spectrometry identification:
The method of mass spectrometry identification is basically the same as the setup in the previous section, but the following points should be noted:
(1) Establishing the spot set
This step is slightly different from the previous section, the mass spectrometry uses a standard sample target of 384, and the LC spot target is a user-defined sample template, so the corresponding parameters should be set according to the actual spot target situation, the number and location of the spots. The following figure:
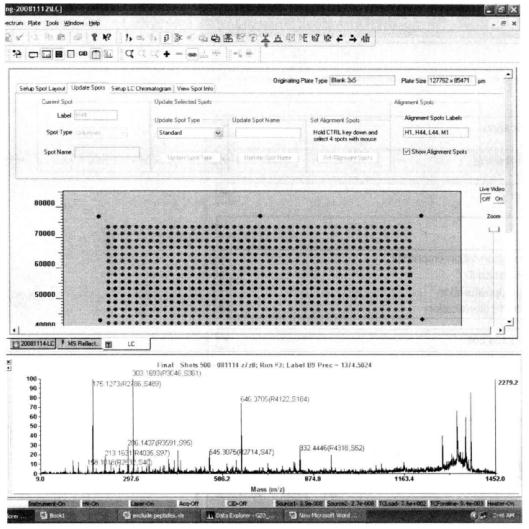
The area surrounded by four red dots is the area of the dot target, which needs to be selected manually.
(2) Acquisition method:
① Firstly, the primary spectrum was obtained by MS reflection mode.
② Automatically select the parent ions for the second level using the Interpretation method, generally 15 to 20 parent ions are selected.
③ Total number of primary MS laser hits: 1000.
④ Total number of secondary MS/MS laser hits: 2000 to 2500.
⑤ Collision energy 2kV, CID off.
(3) Processing method: the same as the previous section.
(4) Run according to the above settings to obtain mass spectrometry data
(5) Mass spectrometry data analysis*8: Use Protein PilotTM software to identify the results obtained by the mass spectrometer.
① Open the software and set up the analysis method as shown in the figure below*9:
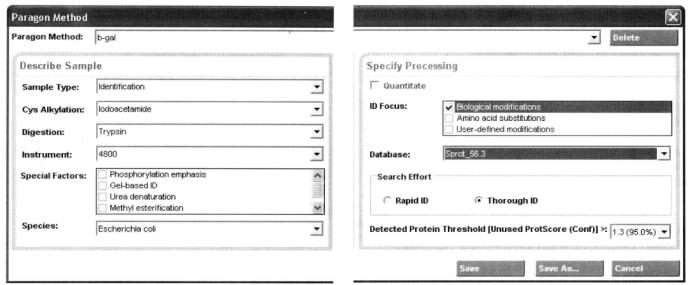
Name and save the file after the method is set up.
② Select the data file to be analyzed:

③ Run the program to obtain the results as shown below:
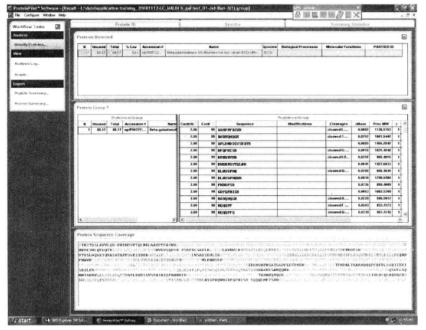
1. The LC must be thoroughly flushed and balanced before use. Before putting on the sample, do a pre-experiment with the standard to check whether the whole system has been flushed clean and meets the requirements for separating the test sample.
2. The reagents selected for LC are all HPLC grade.
3. Protein samples must be purified before LC separation to avoid blocking the column due to impurities in the sample.
*1 The reagents used in the mass spectrometry experiment are all HPLC grade. The water used is ultrapure water.
*2 Determine the amount of trypsin added based on the sample concentration.
*3 When not in use for a long time, some impurities may block the column, so the column should be flushed before use until the pressure and flow rate of the column is stable.
*4 The purpose is to balance the entire liquid phase system, avoid blockages and bubble formation, and stabilize the baseline.
*5 To understand the peak time of the sample, it is necessary to conduct preliminary experiments with excess samples, observe the peak time of the sample, and avoid any samples that have not been collected.
*6 Parameters set based on the peak time of total protein in Leymus chinensis.
*7 LC has good compatibility with automatic sampling device and mass spectrometer.
*8 GPS ExplorerTM can also perform data analysis on LC-separated samples, and usually Protein PilotTM is widely used.
*9 Set sample type, cysteine alkylation, protein digestion enzymes, special factors (optional), species type of sample, selected database, and reliability.

