Principle and Protocol of Expression and Purification of MBP Fusion Protein
Maltose binding protein (MBP) is a periplasmic protein encoded by E. coli mal E gene, which is a member of the bacterial maltose transport system. MBP can bind maltose at micromolar level, so the specific binding of MBP to maltose can be used to purify the fusion protein.
To understand the basic principle of purification of maltose binding protein (MBP) fusion protein. Master the expression and purification methods and main operation steps of MBP fusion protein.
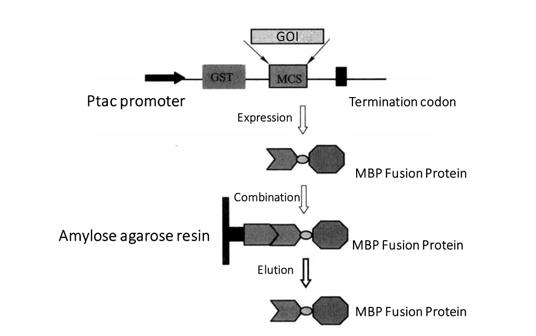 Figure 3-4-1 MBP Fusion Protein Purification Process
Figure 3-4-1 MBP Fusion Protein Purification Process
This section mainly introduces the expression and purification of pMAL-c2 vector and MBP fusion protein. pMAL series vectors are the expression vectors of MBP fusion proteins. Each vector of pMAL series has E. coli mal E gene, encoding maltose binding protein, followed by polyclonal site and termination codon (Figure 3-4-1). Lac I gene encodes lac worker repressor protein, which binds to the Ptac promoter to repress the expression of MBP fusion protein. After IPTG induction, the MBP fusion protein was expressed by the disinhibitory effect. The foreign proteins expressed by pMAL series vectors are fused with MBP, so they can be purified by agarose affinity chromatography resin solidified with maltose.
1. Main Instruments and Equipment
Pipette, micro centrifuge, 1.5mL centrifuge tube, 50mL centrifuge tube, thermostatic water bath, ice maker, thermostatic shaker, ultra clean workbench, thermostatic incubator, shaker, oscillator, centrifuge, metal bath, chromatographic column.
2. Experimental Materials
Vector: pMAL-c2
E. coli strain: BL21 (DE3)
3. Main Reagents
(1) 0.3 mol/L IPTG.
(2) SDS, arc, bis.
(3) Amylose agarose resin, chromatographic column.
(4) Rich medium (containing glucose and ampicillin)
| 10 g/L | Tryptone |
| 5g/L | Yeast extract |
| 5g/L | NaCl |
| 2g/L | Glucose |
| Autoclaved (121°C, 0.105 MPa, 20 min) with ampicillin (100 μg/L). | |
(5) Column washing buffer
| 20 mmol/L | Tris-HCl |
| 200 mmol/L | NaCl |
| 1 mmol/L | EDTA |
| 1 mmol/L | DTT |
(6) Maltose elution buffer
| 10 mmol/L | Maltose |
| 20 mmol/L | Tris-HCl |
| 200 mmol/L | NaCl |
| 1 mmol/L | EDTA |
| 1 mmol/L | DTT |
1. Induced Expression of Target Proteins
(1) The constructed MBP fusion expression plasmid was transformed into BL21 (DE3) strain *1.
(2) Single colonies were picked and inserted into 5mL of Rich medium and incubated overnight at 37°C.
(3) Take 50μL of overnight culture into 5mL of Rich medium and incubate with shaking at 37°C for more than 2h to mid-log growth (A600=0.5-0.6).
(4) Aspirate 1 mL of uninduced culture in a microcentrifuge tube and process as described in steps (6) and (7) below, leaving 1 mL of the culture ready for expansion.
(5) IPTG was added to the remaining culture to a final concentration of 0.3 mmol/L and the culture was continued at 37°C with aeration.
(6) After 3h of induction, 500μL of the sample was placed in a microcentrifuge tube and centrifuged at high speed for 1min at room temperature.
(7) The precipitate was suspended in 100μL 1 × SDS gel spiking buffer, heated at 100°C for 5min, centrifuged at high speed at room temperature for 1min, placed on ice, and all samples were processed and uploaded.
SDS-PAGE*2 electrophoresis.
Coomassie brilliant blue staining showed the position (relative molecular weight) of the expressed product bands.
2. Extended Cultivation
(1) After Coomassie brilliant blue staining to determine the expression of the target protein, the retained bacterial broth was subjected to expanded culture.
(2) Take 0.8mL of retained bacterial broth into 80mL LB culture, and incubate in a 1L shake flask with shaking at 37°C. for more than 2h to mid logarithmic growth (A600=0.5-0.6).
(3) Add IPTG to the culture to a final concentration of 0.3 mmol/L and induce the expression of the target protein at the optimal time and temperature determined by the pre-experiment.
(4) After induction for the appropriate time, the cells were collected by centrifugation at 4000g for 20 min at 4°C.
3. Preparation of Crude Extract of Bacterial Protein
(1) The cell precipitation of 40mL cell culture was suspended in 5mL column washing buffer.
(2) Put the cells in an ice bath, and crush the cells with an ultrasound instrument *3.
(3) Put the centrifuge tube into the freezing centrifuge, adjust the temperature to 4°C, rotate at 9000g, centrifuge for 20 min, remove the cell fragments, and transfer the supernatant into a new tube.
4. Protein Purification and Detection
(1) Rinse the chromatography column 3 times with 2 mL of deionized water, and equilibrate the column 3 times with 2 mL of column washing buffer.
(2) Invert and mix the straight-chain starch-agarose resin, and take 200 μL into the chromatography column.
(3) Resuspend the agarose 3 times with 2 mL of column washing buffer.
(4) The supernatant after centrifugation was slowly added to the chromatography column at 4°C and combined with shaking for 30 min.
(5) Rinse the straight-chain starch-agarose resin 3 times with 600 μL column washing buffer and collect the effluent.
(6) The protein was eluted with 250 μL maltose 3 times and the effluent were collected.
(7) Coomassie brilliant blue staining test (Figure 3-4-2).
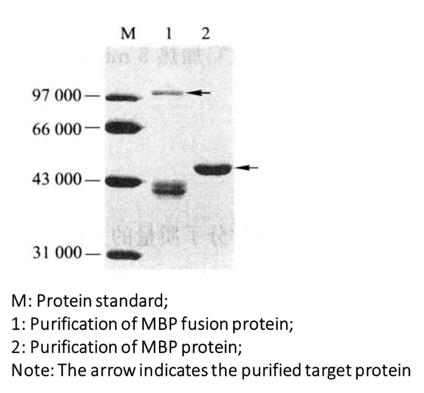 Figure 3-4-2 Purified Gel Electrophoresis of MBP Fusion Protein
Figure 3-4-2 Purified Gel Electrophoresis of MBP Fusion Protein
*1 Or other expression strains.
*2 The concentration of gel is determined according to the relative molecular weight of the target protein.
*3 Cell lysis is usually broken by ultrasound or lysozyme.

