Principle and Protocol of Isotope Labeling
Proteomics is used to study the association of complex protein groups in organisms, involving protein function and protein interaction analysis. Functional analysis is usually used to study the changes of protein group composition under different physiological conditions. Its technical means mainly include protein separation and protein identification. Two-dimensional gel electrophoresis is difficult to separate hydrophobic proteins, proteins with very high or low molecular weight, proteins with extremely acidic or alkaline isoelectric points, and proteins with very low content, For these proteins, the use of isotope tags for relative and absolute quantification (iTRAQ) based on liquid chromatography and mass spectrometry can effectively overcome the shortcomings of reverse gel electrophoresis. In addition, iTRAQ technology has some unique advantages over two-dimensional gel electrophoresis technology:
① It can distinguish more proteins in the state of post-transcriptional modification;
② Multiple groups of samples can be compared at the same time. At present, at most 8 groups of samples can be compared at the same time under the same separation conditions;
③ Absolute or relative quantification can be performed for specific target proteins;
④ Because it can analyze multiple groups of samples at the same time, it increases the correlation of experimental statistics.
The disadvantage of TRAQ technology is that it can only use mass spectrometry technology to identify the peptides and isotope tags that have been enzymatically hydrolyzed. The obtained data need to undergo a lot of calculations and match with the known sequences and modification types in the database, so it is not easy to use for new species or new proteins that lack database support.
The purpose of this experiment guide is to help researchers master the basic principles of iTRAQ technology and understand the experimental process and precautions of iTRAQ.
ITRAQ reagent currently has 8 kinds of isotope labels. The structure of the labels is shown in Figure 6-3-1. These labels can be specifically combined with the N-terminal of the peptide segment. In the primary mass spectrometry mode, the same peptide segment labeled by different labels has the same mass number, while in the secondary mass spectrometry mode, each label can produce corresponding strong signal characteristic ions, The purpose of sample quantitative analysis can be achieved through the quantitative analysis of the characteristic ion strength of the label. The iTRAQ experimental process is shown in Figure 6-3-2.
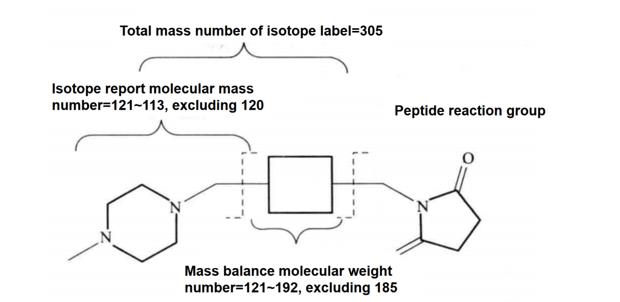 Figure 6-3-1 iTRAQ isotope label structure
Figure 6-3-1 iTRAQ isotope label structure
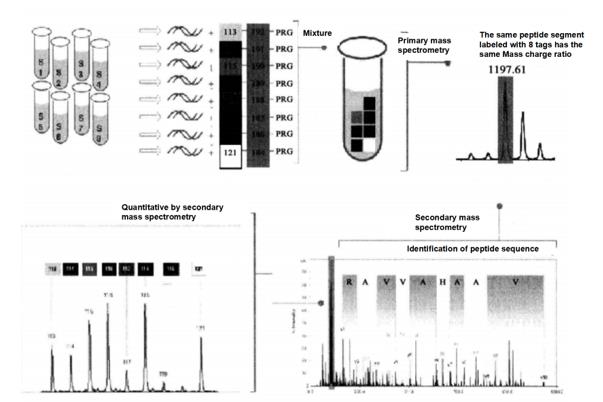 Figure 6-3-2 Overview of iTRAQ experiment process
Figure 6-3-2 Overview of iTRAQ experiment process
1. Main Instruments and Equipment
Micro-pipette, vortex oscillator, centrifuge, constant temperature water bath, high performance liquid chromatography, mass spectrometer.
2. Material
Prepared protein sample.
3. Main reagents
(1) iTRAQ®Reagents-8plex One Assay or Multi-Plex Kits:
Isotope labeling solution 113 (iTRAQTMreagent 113);
Isotope labeling solution 114 (iTRAQTMreagent 114);
Isotope labeling solution 115 (iTRAQTMreagent 115);
Isotope labeling solution 116 (iTRAQTMreagent 116);
Isotope labeling solution 117 (iTRAQTMreagent 117);
Isotope labeling solution 118 (iTRAQTMreagent 118);
Isotope labeling solution 119 (iTRAQTMreagent 119);
Isotope labeling solution 121 (iTRAQTMreagent 121);
Denaturant solution: 2%SDS;
Reducing reagent;
Cysteine blocking reagent.
(2) Pancreatin.
(3) Reagents for cation exchange column purification:
Cation exchange buffer-load;
Cation exchange buffer-clean; Cation exchange buffer-elute.(4) Liquid chromatography loading buffer:
| Acetonitrile | 2% |
| TFA | 0.1% |
1. Reducing protein and blocking cysteine
(1) Add 20μL of Lysis Buffer and 1μL of Denaturant to each sample tube*1.
(2) Vortex shake and mix well.
(3) Add 2μLof Reducing Reagent to each sample tube.
(4) Mix with vortex shaking and centrifuge briefly.
(5) Hold in a 60°C water bath for 1h.
(6) Centrifuge briefly.
(7) Add 1μLof cysteine blocker to each sample tube.
(8) Mix with vortex shaking and centrifuge briefly.
(9) Leave at room temperature for 10 min.
2. Trypsin digest protein
(1) Add 25μL of ultrapure water to each new trypsin vial to dissolve trypsin*2.
(2) Vortex shake to fully dissolve the trypsin and centrifuge briefly.
(3) Add 2 to 10μL of trypsin solution to each sample tube.
(4) Mix with vortex shaking and centrifuge briefly.
(5) Keep warm overnight (12-16h) at 37°C.
3. iTRAQ® reagents label tryptic digestion products
(1) Equipbrate the iTRAQ® reagent to room temperature.
(2) Centrifuge briefly to concentrate the solution at the bottom of the tube.
(3) Add 50μL of isopropanol to each tube of iTRAQ reagent.
(4) Mix by vortex shaking and centrifuge briefly.
(5) Add each tube of iTRAQ Reagent to the corresponding sample to be labeled.
(6) Vortex and mix, centrifuge briefly.
(7) If the pH of the mixed solution is <7.5, add 5μL of Lysis Buffer.
(8) Allow to stand at room temperature for 2h.
(9) Mix labeled samples into one tube, vortex and mix well, centrifuge briefly*3.
(2) Prepare standard protein BSA control 10μL, control 2μL, sample treated 10μL and treated 2μL, respectively.
4. Purification of samples with cation exchange columns
(1) The sample mixture was diluted more than 10 times with cation exchange column loading buffer to make the salt ion concentration in the solution less than 20 mmol/L and reduce the concentration of organic matter.
(2) The mixture was mixed by vortex shaking.
(3) Remove a small portion of the sample and check if the pH of the solution is between 2.5 and 3.3. If it is not in this range, adjust the pH to 2.5-3.3 with cation exchange column loading buffer or with 1 mol/L phosphoric acid.
(4) Add 1 mL of wash buffer to the cation exchange column and discard the effluent.
(5) Add 2 mL of sample buffer to the cation exchange column and discard the effluent*3.
(6) Slowly drop the diluted sample solution into the cation exchange column (approximately 1 drop/s), reserving the effluent to a new sample tube.
(7) Add 1 mL of loading buffer to wash off TCEP, SDS, calcium chloride and excess iTRAQ reagent from the cation exchange column. Retain the effluent into a new sample tube.
(8) Slowly add 500μL of elution buffer dropwise (approximately 1 drop/s) and collect the eluate into a new sample tube, sample purification is complete.
(9) The purified sample obtained above is vacuum dried and the sample is dissolved with 10μL of pquid chromatography loading buffer. The pquid phase separation of iTRAQ-labeled peptides is performed in the same way as the normal pquid phase method.
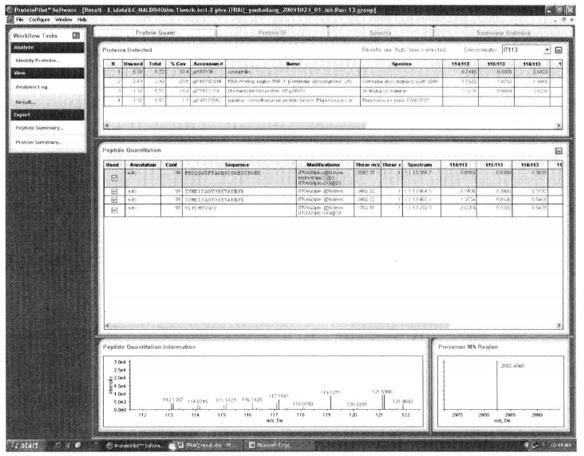
5. Result of iTRAQ
The analysis of the mass spectrometry data by ProteinPilotTM software gives the following information, the sample-to-sample ratio (e.g., 114:113, the ratio of the sample labeled 114 to the sample labeled 113, indicating the difference between the samples), the sequence number of the differential protein, the name of the differential protein and the corresponding species information.
1. Mixing compounds containing sulfhydryl groups (e.g. DTT and β-mercaptoethanol) can affect the negative break of cysteine.
2. High concentrations of detergents or denaturants can affect trypsin activity.
3. Compounds containing primary amine groups (e.g., ammonium acetate, ammonium carbonate, ammonium citrate, ammonium tartrate, ethanolamine, and Tris buffer) can react with iTRAQTM reagents and affect labepng.
*1 Each sample tube contains 5 to 100 μg of protein sample.
*2 Contaminants that may inhibit trypsin digestion activity can be removed by acetone precipitation prior to trypsin digestion.
*3 The labeled samples can be kept at -20°C for several days.

