Principle and Protocol of Yeast Two Hybrid System
The yeast two hybrid system was established by Fields et al. in 1989 based on their understanding of eukaryotic regulation of transcription initiation. Transcription of cell initiation genes requires the participation of trans transcriptional activators, which are modular in structure, that is, these factors are often composed of two or more mutually independent domains, including DNA binding domain (BD) and transcription activating domain (AD). Although BD alone can bind to the promoter, it cannot activate transcription. The transcriptional activation of eukaryotic gene transcription activating factors is jointly completed by BD and AD, which have relatively independent functions. The spatial connection between these two domains through covalent or non-covalent connection makes them close to each other and starts the expression of downstream reporter genes. According to the characteristics of transcriptional activator, the "bait" protein is fused with BD, and the protein that may interact with it or the screened library is fused with AD. If they interact, AD and BD can be spatially close to each other and start the expression of downstream reporter genes.
This classical yeast two hybrid system is widely used in library screening, protein interaction identification and protein interaction domain identification. The yeast two hybrid system uses in vivo experiments to study protein interactions, which can simulate the action environment in eukaryotic cells and is easy to operate. All operations are performed at the nucleic acid level, without the need to purify a large number of proteins, and the weak interaction of proteins can be accurately determined. The "bait" protein in the yeast two hybrid system can be either a complete protein or a functional region of the protein, which further expands its application scope.
Grasp the principle and application of the classic yeast two hybrid system, learn the methods of preparation and transformation of yeast cell competent state, and be familiar with the operation process and experimental precautions of yeast two hybrid system for detecting protein interaction.
The principle of the classical yeast two hybrid system is that the transcriptional activation factors are modular in structure, that is, these factors are often composed of two or more mutually independent domains, including DNA binding domain (BD) and DNA activation domain (AD), which are necessary for transcriptional activation factors to perform their functions. Although BD alone can bind to the promoter, it cannot activate transcription. However, the hybrid proteins formed by BD and AD with different transcription activating factors still have the normal function of activating transcription. According to the characteristics of transcription factors, BD was fused with known bait protein X to construct BD-X plasmid vector. The AD gene is fused with the cDNA library, gene fragment or gene mutant (represented by Y) to construct the AD-Y plasmid vector. Two shuttle plasmid vectors were co transformed into yeast for expression. The interaction of protein X and Y leads to the spatial proximity of BD and AD, thus activating the expression of yeast strain specific reporter genes (such as LacZ, HIS3, LEU2) regulated by the UAS downstream promoter (Figure 4-1-1). Because of the expression of HIS3 or LEU2, the transformants can grow on specific defective media, and because of the expression of LacZ, the transformants show blue in the presence of X-β-Gal.
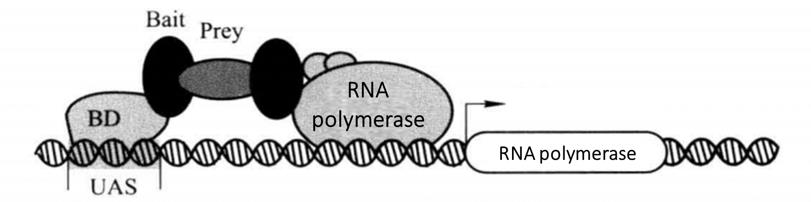 Figure 4-1-1 Schematic Diagram of Classical Yeast Two Hybrid Principle
Figure 4-1-1 Schematic Diagram of Classical Yeast Two Hybrid Principle
1. Main Instruments and Equipment
Micropipette (2 μL, 10 μL, 20 μL, 200 μL, 1000μL), low temperature centrifuge, desktop centrifuge, agarose gel electrophoresis system, protein gel electrophoresis system, gel imaging system, semi dry transfer system, thermostatic shaker, thermostatic incubator, fume hood, ice maker, oscillator, thermostatic metal bath, sterile inoculation ring, 10 cm culture dish, 15 cm culture dish.
2. Experimental Materials
AH109 or Y187 yeast strain*1, yeast expression vector*2.
3. Main Reagents
(1) Various basic media and nutrient deficiency media:
Minimal SD base,minimal SD agar base, YPD medium, YPD agar medium, -Leu DO supplement, -Trp DO supplement, -Leu / -Trp DO supplement, -Leu /-Trp / -His DO supplement, -Leu / -Trp / -His / -Ade DO supplement, adenine, PEG8000,Carring DNA, DMSO, TE / LiAC buffur, PEG / LiAC, X-β-gal.
Na2HPO4·7H2O
NaH2PO4·H2O
KCl
MgSO4·7H2O
(2) LB medium, Carboxybenzylpenicillin and Kanamycin.
(3) Cracking buffer
| 8 mol/ | Urea |
| 5% | SDS |
| 40 mmol/L | Tris-HCl (pH 6.8) |
| 0.1 mmol/L | EDTA |
| 0.4 mg/ml | Bromophenol blue |
(4) Z-buffer (pH 7.0)
| 16.1 g/L | Na2HPO4·7H2O |
| 5.5 g/L | NaH2PO4·H2O |
| 0.75 g/L | KCl |
| 0.246 g/L | MgSO4·7H2O |
(5) Z-buffer / X-β-gal solution
| 100 mol/L | Z-buffer |
| 0.27 mol/L | β-Mercaptoethanol |
| 1.67 mol/L | X-β-Gal storage solution |
1. Resuscitation and Phenotype Verification of Yeast
(1) 1 to 2 days before resuscitation, YPDA agar was prepared and poured after high-pressure disinfection.
(2) Pick up a small group of yeast cells from the yeast cryopreservation tube with a sterile inoculum ring, and inoculate them onto the YPDA agar plate with repeated zigzag method.
(3) Incubate upside down at 30°C for 3-5 days.
2. Plasmid Transformation into Yeast Cells (Conventional Transformation)
(1) Yeast clones with a diameter of 2-3 mm were inoculated into 0.5 mL YPDA medium.
(2) The cell clots were evenly dispersed by violent vibration.
(3) Transfer the cells into a conical flask containing fresh YPDA medium *3.
(4) At 30°C, 250r/min, incubate for 16-18h by shaking until the stable period (A600 >1.5).
(5) Transfer the overnight culture into a conical flask containing fresh YPDA medium *4, and A600 0.2-0.3.
(6) 30°C, 230-270r/min, shake incubation to make A600 reach 0.5±0.1 (generally 3h).
(7) The cells were transferred to several 50mL centrifuge tubes, 1000g, and centrifuged at room temperature for 5min.
(8) Discard the supernatant, add 25-50 mL of sterilized H2O, shake and resuspension, wash and collect cells.
(9) 1000g, centrifugate at room temperature for 5min, and discard the supernatant.
(10) Add 1mL fresh sterile 1 × TE / LiAC suspension cells *5.
(11) Prepare 1.5mL sterile EP tubes and add 0.1 μg BD/X, AD/Y transfected plasmid, 0.1 mg vector DNA into each tube.
(12) Add competent cells resuspended by 1 × TE / LiAC and mix gently*6.
(13) Add an appropriate volume of 1×PEG/LiAC to each tube and mix with high-speed shaking*7.
(14) Incubate at 30°C, 200r/min, with shaking for 0.5h.
(15) Add an appropriate volume of DMSO*8 and mix by gentle inversion without shaking.
(16) Heat shock in a 42°C water bath for 15min. For large conversions, shake frequently to mix.
(17) Place on ice for 5-10min and centrifuge cells at 12000g for 5s at room temperature.
(18) Remove the supernatant and add the appropriate volume of 1×TE to resuspend the cells according to the number of plates laid*9.
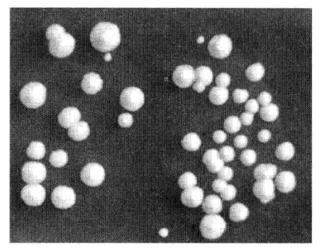 Figure 4-1-2 Results of Yeast Transformation
Figure 4-1-2 Results of Yeast Transformation
(19) Spread the plates and invert the plates and incubate at 30°C until clones appear (Figure 4-1-2).
3. Detection of Target Protein Expression in Yeast Cells
(1) A BD/X fresh yeast clone of approximately 2 mm in diameter containing the target protein gene fragment was picked in 5 mL of the corresponding nutrient-deficient liquid medium and incubated for 16 h*10 at 30°C, 230 r/min with shaking.
(2) The overnight culture was expanded in 50mLYPDA medium at 30°C, 220-250r/min to achieve A600 values of 0.4-0.6*11.
(3) Centrifuge at 1000g for 5min and discard the supernatant.
(4) Add 30 mL of H2O and resuspend the cells. 1000g, centrifuge for 5 min, discard the supernatant, and add liquid nitrogen for quick-freezing precipitation*12.
(5) After the liquid nitrogen evaporates completely, add lysis buffer at a ratio of 100μL/7.5 A600 to resuspend the cells and transfer to a 1.5mL EP tube.
(6) Add acidic glass beads to the EP tube at a ratio of 80 μL/7.5 A600. heat at 70°C for 10 min and shake vigorously for 1 min.
(7) Centrifuge at 12000g for 5min at 4°C. Transfer the supernatant to a new 1.5mLEP tube on ice.
(8) Heat in a water bath at 100°C for 3-5min and shake vigorously for 1min.
(9) Centrifuge at 12000g for 5min at 4°C. Combine the two supernatants into one EP tube.
(10) Take 20μL of the supernatant, add the loading buffer, and denature at 100°C for 5min.
(11) After SDS-PAGE electrophoresis, the expression of the target protein in yeast cells was detected by immunoblotting technique.
4. Detection of β-galactosidase Secretion by Filter Paper Transfer Assay
(1) When the yeast has grown to a diameter of 1.5-3 mm in size, sterile, appropriately sized Whatman filter paper is taken and applied to the yeast plate.
(2) After the filter paper was largely wetted by the water on the agar plate, the filter paper was removed, at which point the yeast clones were transferred to the filter paper.
(3) The filter paper with the yeast clones attached was snap frozen in liquid nitrogen for more than 13 s.
(4) The freshly prepared Z-buffer/X-β-Gal liquid is dropped onto another sterile Whatman filter paper and the filter paper is completely wetted*13.
(5) The filter paper with the yeast clone attached is carefully attached to the filter paper soaked with Z-buffer/X-β-Gal.
(6) Incubate at room temperature for 8-16h and wait for the appearance of blue spots (Figure 4-1-3) *14.
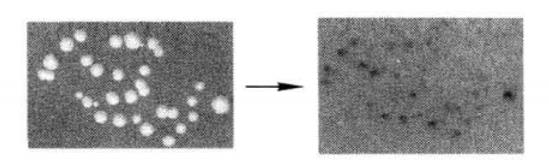 Figure 4-1-3 Detection of β-galactosidase Expression by Filter Paper Transfer Assay
Figure 4-1-3 Detection of β-galactosidase Expression by Filter Paper Transfer Assay
Setting of yeast two-hybrid control
A positive control, a negative control, and a chromogenic system control are generally set up for routine yeast two-hybrid operations, using the MATCHMAKER GAL4 yeast two-hybrid screening system as an example:
(1) Positive control: pGADT7-T + pGBKT7-53, where T protein and 53 protein are two proteins that have been shown to bind and initiate reporter gene expression in yeast cells.
(2) Negative control: pGADT7-T + pGBKT7-lam, where it is clear that the T and lam proteins are unable to bind in yeast cells.
(3) Systematic chromogenic control: pGADT7-Pcll, where this expression plasmid can cause the secretion of β-galactosidase and when transferred into yeast cells, thus testing whether there is a problem with the chromogenic system.
1. Common problems and reference solutions
(1) Transformation efficiency is too low. Try to use fresh medium as well as fresh and 2~3mm diameter yeast gram drop to ensure the yeast is alive. The plasmids used for transformation can be ethanol precipitated before use to improve the purity and concentration of the plasmids.
(2) High background: When HIS3 is used as a reporter gene, the background may be too high because the HTS3 gene has a certain degree of leaky expression, and then the appropriate amount of HIS3 protein competitive inhibitor 3-AT (3-amino-1,2,4-triazole) can be used to reduce the background. Alternatively, a more stringent screen for reporter genes such as Ade can be added.
(3) The "bait" protein has the phenomenon of self-activation: the method of cloning mutation can be used to knock out or mutate a segment of amino acid sequence that generates self-activation, but this method may destroy the interaction between proteins.
*1 Different yeast two-hybrid systems have different yeast strains to choose from.
*2 Commercially available yeast two-hybrid systems generally provide the appropriate companion yeast expression plasmids and positive and negative control plasmids. Here we use the pGBKT7 and pGADT7 yeast expression plasmids provided by the Matchmaker GAL4 two-hybrid system.
*3 The amount of YPDA used depends on the size of the transformation. 25 mL YPDA is recommended for small transformations and 50 mL YPDA for large transformations. In addition, the volume of the conical flask should be 3-5 times the combined volume of the added medium and yeast cells in order to work well for amplifying yeast cells.
*4 The amount of YPDA used depends on the scale of transformation. 150mL YPDA is recommended for small transformations and 300mLYPDA for large transformations.
*5 The volume of 1×TE/LiAC depends on the number of plates laid and the size of the plates. Usually, 1×TE/LiAC can be added at a ratio of 100 μL. per 1 10 mm Petri dish.
*6 The volume of receptor cells added to each tube depends on the number of yeast cells and the size of the plate, usually 100 μL of receptor cells per tube for small transformations and 1 mL of receptor cells per tube for large transformations. If the clones are too dense after growth, the volume of receptor cells added in each tube can be reduced appropriately, and vice versa, the volume of receptor cells can be increased appropriately.
*7 Add 600 μL per tube for small transformations; add 6 ml per tube for large transformations.
*8 Add 70 μL per tube for small transformations; 700 μL per tube for large transformations.
*9 Generally use 100 μL volume plates for 100 mm dishes. 200 μL volume plates for 150 mm dishes.
*10 Which nutrient defect should be determined by the reporter gene of the vector contained in the yeast clone.
*11 Total A value of the sample = measured A value × total volume of the sample (mL).
*12 The precipitate can be placed into -80°C for long-term storage after snap-freezing in liquid nitrogen.
*13 The amount of Z-buffer/X-β-Gal should be dropped in such a way that it just wets the filter paper. If the amount of liquid is too much, it tends to diffuse the yeast cell surface-galactosidase. Generally, a Whatman filter paper of about 9 cm should be filled with about 1 mL of Z-buffer/X-β-Gal.
*14 The time of appearance of blue spots varies depending on the strength of the binding between the proteins being tested. Generally, the stronger the binding between the proteins, the earlier and darker the blue spots appear. In addition, various two-hybrid systems have a maximum color development time limit, and any blue spots appearing after this time are not experimentally significant.

