H4 Family
Related Symbol Search List
- HIST1H4A
- HIST1H4B
- HIST1H4C
- HIST1H4D
- HIST1H4E
- HIST1H4F
- HIST1H4G
- HIST1H4H
- HIST1H4I
- HIST1H4J
- HIST1H4K
- HIST1H4L
- HIST4H4
Immunology Background
Background
Histone H4 is a core histone protein that plays a crucial role in the organization and packaging of DNA into a compact structure called chromatin. It is a member of the histone H4 family, which consists of several closely related variants. The histone H4 family is involved in regulating gene expression and modulating chromatin structure to facilitate various cellular processes.
Structure
Histone H4 is an 11.3 kDa protein that is made up of 102 amino acid residues. It is one of the core histones, along with H2A, H2B, and H3, and is considered to be the most conserved histone. Core histones are among the most evolutionarily conserved proteins in the eukaryotic nucleus. It has a highly conserved globular core domain, which forms the histone fold. The histone fold consists of three α-helices (α1, α2, and α3) connected by two loops (L1 and L2). The histone fold is responsible for the assembly of the nucleosome, the basic repeating unit of chromatin. At the N- and C-termini of histone H4, there are flexible regions known as the N-terminal tail and the C-terminal tail, respectively. These tails can undergo various post-translational modifications, such as acetylation, methylation, and phosphorylation, which regulate chromatin structure and gene expression.
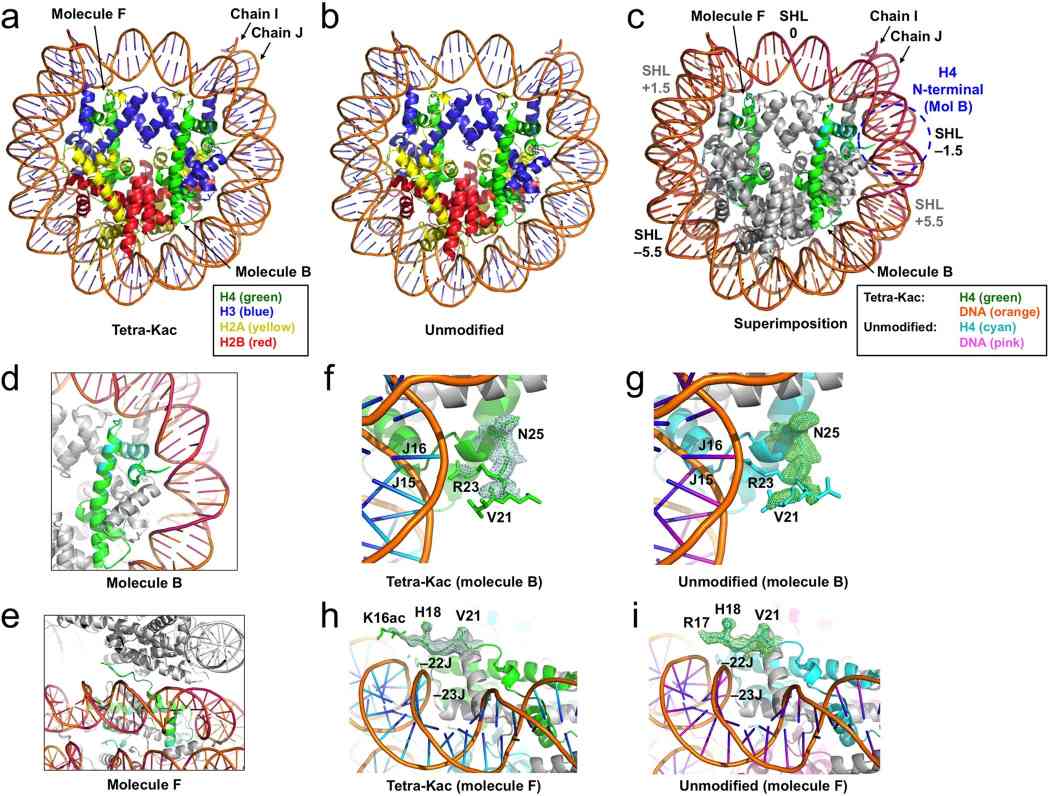 Fig.1 Crystal structures of the H4-tetra-acetylated nucleosome core particle (NCP). (Wakamori M, et al., 2015)
Fig.1 Crystal structures of the H4-tetra-acetylated nucleosome core particle (NCP). (Wakamori M, et al., 2015)Function
The histone H4 family is essential for the organization, stability, and regulation of chromatin structure, thereby influencing various cellular processes, including gene expression, DNA repair, and development.
| Functions | Details |
|---|---|
| Nucleosome Formation | Histone H4 plays a crucial role in nucleosome formation, which is the primary mechanism of DNA compaction in eukaryotic cells. Together with histones H2A, H2B, and H3, it forms the core histone octamer around which DNA is wrapped to form a nucleosome. |
| Chromatin Structure and Stability | Histone H4, along with other core histones, helps maintain the structure and stability of chromatin. The N-terminal tail of histone H4 interacts with neighboring nucleosomes and other nuclear proteins, contributing to higher-order chromatin organization. |
| Epigenetic Regulation | The histone H4 family members, including its variants, are involved in epigenetic regulation. Post-translational modifications of histone H4, such as acetylation and methylation, can influence chromatin structure and gene expression. These modifications can recruit various effector proteins that either activate or repress gene transcription, ultimately influencing cellular processes. |
| DNA Repair | Histone H4 also plays a role in DNA repair mechanisms. It can undergo modifications in response to DNA damage, facilitating the recruitment of repair factors to damaged sites and promoting efficient DNA repair processes. |
| Development and Differentiation | Different variants of histone H4 are expressed during specific stages of development and cellular differentiation. They contribute to the regulation of gene expression patterns required for cell fate determination and tissue-specific functions. |
The histone H4 family and its variants play crucial roles in cell cycle regulation, gene expression regulation, and chromatin structure. Here's a discussion of their important functions in these processes:
| Functions | Details |
|---|---|
| Cell Cycle Regulation |
|
| Gene Expression Regulation |
|
| Chromatin Structure |
|
In summary, the histone H4 family and its variants play crucial roles in cell cycle regulation, gene expression regulation, and chromatin structure. Their contributions to nucleosome assembly, histone modifications, chromatin remodeling, and higher-order chromatin organization are essential for proper cell function, development, and the maintenance of genomic stability.
Members and Variants of the H4 Family
The histone H4 family has 116 members found in different organisms. Some examples of histone H4 variants include: HIST4H4, HIST2H4B, HIST1H4I, HIST1H4A, HIST1H4D, HIST1H4F, HIST1H4K, HIST1H4J, HIST1H4C, and HIST1H4H.
| Members and variants | Details |
|---|---|
| Canonical Histone H4 | This is the standard form of histone H4 found in nucleosomes. It is involved in the packaging of DNA into chromatin and provides structural support to the nucleosome core particle. |
| Histone H4 Variants |
HIST1H4A (Histone H4A): This variant of histone H4 is one of the core histones involved in nucleosome assembly and stability. It is associated with gene regulation and chromatin structure modulation. HIST1H4A is expressed in a wide range of tissues and is essential for maintaining the integrity of chromatin. HIST1H4B (Histone H4B): HIST1H4B is a highly conserved variant of histone H4 and plays a crucial role in chromatin packaging and stability. It contributes to the maintenance of chromatin structure and is involved in gene expression regulation. HIST1H4B is expressed in various tissues and is essential for proper chromatin organization. HIST1H4C (Histone H4C): It participates in nucleosome formation and chromatin compaction. It is involved in regulating gene expression and contributes to chromatin organization and stability. HIST1H4D (Histone H4D): It is associated with gene expression regulation and chromatin remodeling. HIST1H4D contributes to the dynamic modulation of chromatin structure, allowing for proper gene regulation and cellular processes. HIST1H4E (Histone H4E): It is involved in chromatin organization and epigenetic regulation. This variant of histone H4 contributes to the modulation of gene expression patterns and is important for cellular development and differentiation. HIST1H4F (Histone H4F): It plays a role in gene regulation and is involved in chromatin structure modulation. HIST1H4F is expressed in various tissues and contributes to the dynamic control of gene expression. HIST1H4G (Histone H4G): It is essential for nucleosome assembly and contributes to the maintenance of chromatin integrity. It plays a crucial role in chromatin organization and stability, ensuring proper gene regulation and cellular function. HIST1H4H (Histone H4H): It is involved in higher-order chromatin organization and compaction. HIST1H4H contributes to the formation of chromatin domains and helps maintain the overall structure of chromatin. HIST1H4I (Histone H4I): HIST1H4I participates in epigenetic regulation and control of gene expression. It is involved in chromatin remodeling and contributes to the dynamic modulation of chromatin structure to facilitate proper gene regulation. HIST1H4J (Histone H4J): It is associated with nucleosome stability and maintenance of chromatin structure. HIST1H4J contributes to the organization and integrity of chromatin, ensuring proper gene expression and cellular processes. HIST1H4K (Histone H4K): It is involved in chromatin remodeling and modulation of gene expression patterns. It contributes to the dynamic control of chromatin structure and plays a role in regulating gene activity. HIST1H4L (Histone H4L): It contributes to nucleosome assembly and chromatin compaction. HIST1H4L is involved in maintaining chromatin integrity and ensuring proper gene regulation. |
These members and variants of the histone H4 family have specific functions in regulating chromatin structure, gene expression, and epigenetic processes. By undergoing various post-translational modifications, they provide a dynamic and versatile mechanism for modulating gene activity and maintaining genomic stability.
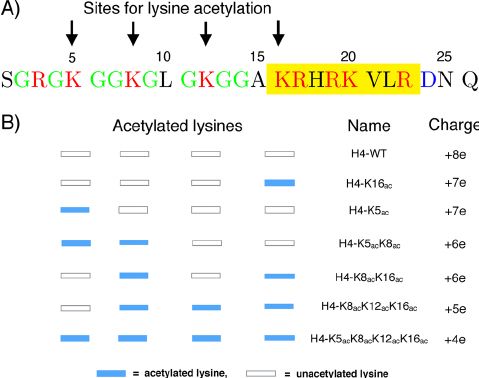 Fig.2 The H4 N-terminal histone tail sequence and acetylation sites. (Winogradoff D, et al., 2015)
Fig.2 The H4 N-terminal histone tail sequence and acetylation sites. (Winogradoff D, et al., 2015)The Role of Histone H4 Family in the Occurrence and Development of Diseases
The histone H4 family and its variants have been implicated in the occurrence and development of various diseases, including cancer and neurodegenerative diseases. Here's an overview of their roles in these conditions:
Cancer
Alterations in histone modifications and chromatin structure are common in cancer cells, leading to dysregulated gene expression and genomic instability. The histone H4 family members and their modifications play crucial roles in these processes. Some specific roles include:
- Histone H4 variants, such as H4K20me3 (histone H4 trimethylated at lysine 20), are associated with gene silencing and have been found to be reduced in several cancer types. Loss of H4K20me3 can lead to the activation of oncogenes and contribute to tumor development.
- Dysregulation of histone acetylation, including H4 acetylation, is commonly observed in cancer. Increased acetylation of histone H4 is associated with transcriptional activation of oncogenes and tumor progression.
- Histone H4 mutations have been identified in specific types of cancer. For example, mutations in the HIST1H4C gene have been found in pediatric brain tumors, known as diffuse intrinsic pontine gliomas (DIPG). These mutations may affect chromatin structure and gene expression, contributing to tumor development.
Neurodegenerative Diseases
Epigenetic dysregulation has been implicated in neurodegenerative diseases, and histone modifications play a critical role in regulating gene expression patterns in the brain. The histone H4 family and its modifications have been implicated in neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and Huntington's disease. Some specific roles include:
- Dysregulation of histone acetylation, including histone H4 acetylation, has been observed in neurodegenerative diseases. Reduced histone acetylation can lead to altered gene expression patterns and contribute to neuronal dysfunction and degeneration.
- Abnormal deposition of modified histones, including histone H4, in the brain has been observed in neurodegenerative diseases. This can affect chromatin structure and gene expression, contributing to disease pathology.
- Histone modifications, including H4 modifications, can influence the expression of genes associated with neurodegenerative disease-related proteins, such as amyloid-beta and tau in Alzheimer's disease, α-synuclein in Parkinson's disease, and mutant huntingtin in Huntington's disease.
- Epigenetic modulation of histone H4 modifications and chromatin structure has been explored as a potential therapeutic approach for neurodegenerative diseases. Targeting histone modifications may help restore normal gene expression patterns and mitigate disease progression.
Overall, the histone H4 family and its modifications play important roles in the occurrence and development of diseases, including cancer and neurodegenerative diseases. Understanding the specific mechanisms underlying these roles can provide insights into disease pathology and potentially lead to the development of novel therapeutic strategies.
Case Study
Case 1: Chou RH, Wang YN, Hsieh YH, et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell. 2014;30(2):224-237.
Histone H4-K20 methylation is critical for DNA synthesis and repair. However, the regulation of these functions by upstream stimuli is not well understood. In this study, researchers identified a tyrosine phosphorylation site at Y72 of histone H4. This phosphorylation site facilitated the recruitment of histone methyltransferases (HMTases) SET8 and SUV4-20H, enhancing the K20 methylation of histone H4. This, in turn, promoted DNA synthesis and repair.
To investigate the impact of H4-Y72 phosphorylation on DNA synthesis, cells were labeled with BrdU, and the newly synthesized DNA associated with histone H4 (wild-type or Y72F mutant) was measured. The results showed that EGF treatment increased the amounts of newly synthesized DNA in wild-type histone H4 but not in the Y72F or K20R mutants. Inhibiting EGF receptor activity attenuated EGF-stimulated DNA synthesis in the wild-type histone H4. The Y72F mutant, which lacked phosphorylation, displayed reduced DNA synthesis ability, similar to the K20R mutant, which lacked methylation. These findings supported the role of H4-Y72 phosphorylation in promoting DNA synthesis.
The researchers also examined the effect of H4-Y72 phosphorylation on cell cycle progression. Cells expressing wild-type or Y72F mutant histone H4 were treated with nocodazole to inhibit mitosis and assess the progression from the G1 to the S phase. The Y72F mutant showed reduced cell cycle progression compared to the wild-type, with fewer cells in the G1 phase. Treatment with AG1478, an inhibitor of EGF receptor activity, abolished the differences, indicating that EGF signaling influences cell cycle progression through H4-Y72 phosphorylation.
Further experiments using synchronized cells revealed that the Y72F mutant entered the S phase more slowly than the wild-type. EGF stimulation increased histone H4-Y72 phosphorylation, K20 methylation, and DNA synthesis in a dose-dependent manner. Cell proliferation assays demonstrated that the Y72F mutant had slower growth compared to the wild-type. Collectively, these results indicated that the Y72F mutant impaired DNA synthesis, delayed S phase progression, and exhibited slower cell growth due to defective K20 methylation.
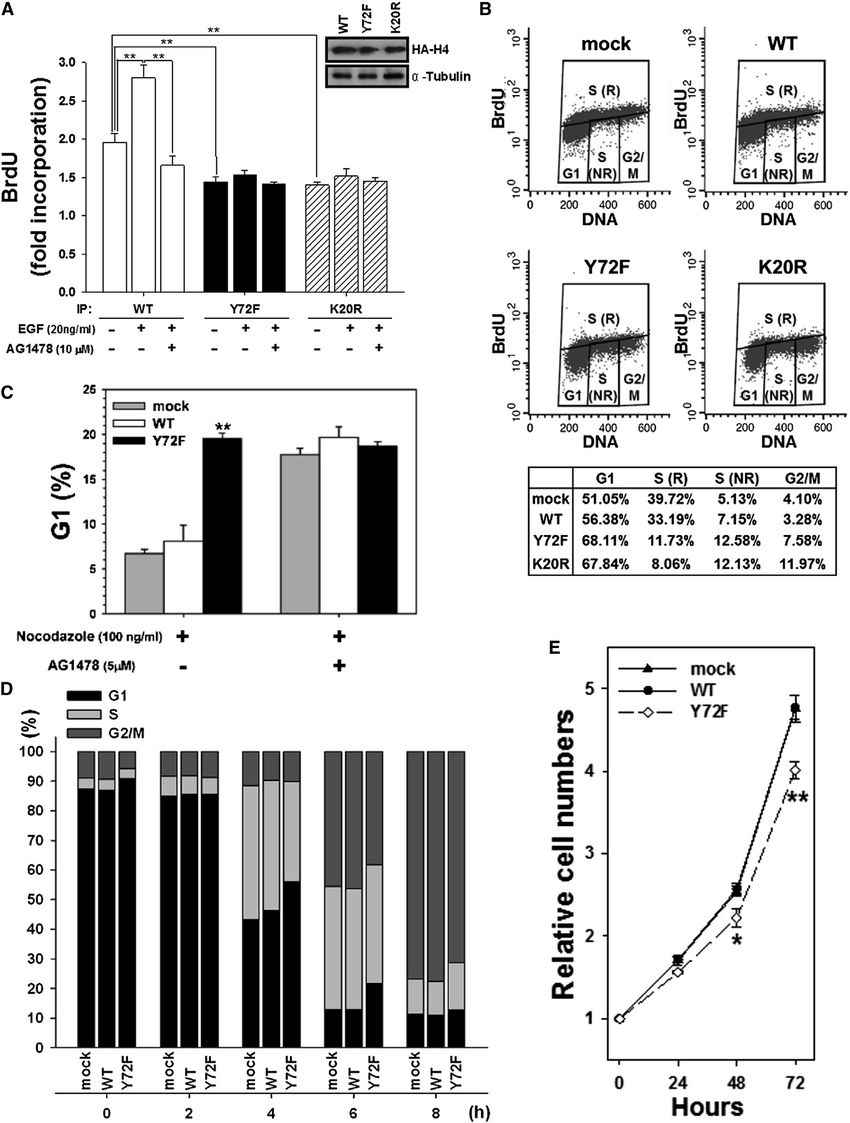 Fig.1 Histone H4-Y72 phosphorylation is involved in DNA synthesis.
Fig.1 Histone H4-Y72 phosphorylation is involved in DNA synthesis.Case 2: Wang R, Zheng X, Zhang L, et al. Histone H4 expression is cooperatively maintained by IKKβ and Akt1 which attenuates cisplatin-induced apoptosis through the DNA-PK/RIP1/IAPs signaling cascade. Sci Rep. 2017;7:41715.
In this study, the researchers investigated the role of histone H4 expression in chemoresistance and the antitumor efficacy of cisplatin. They found that downregulation of histone H4 sensitized lung cancer cells to cisplatin-induced apoptosis and enhanced the cytotoxicity of cisplatin. This effect was associated with the proteasomal degradation of RIP1, accumulation of cellular reactive oxygen species (ROS), and degradation of inhibitor of apoptosis proteins (IAPs). Furthermore, they discovered that histone H4 knockdown suppressed cisplatin-induced DNA-dependent protein kinase (DNA-PK) activation, and inhibiting DNA-PK reduced the expression of RIP1 and IAPs in cisplatin-treated cells.
To validate these findings in vivo, the researchers used a mouse xenograft tumor model. They injected control cells and histone H4 knockdown cell clones into mice and treated them with cisplatin. Consistent with the in vitro results, histone H4 knockdown significantly enhanced the antitumor activity of cisplatin. Downregulation of histone H4 was confirmed in the xenograft tumor tissues, and the levels of RIP1 and IAPs were decreased after cisplatin treatment in the histone H4 knockdown groups. TUNEL assay revealed increased apoptotic cell numbers in the tumor tissues from the cisplatin-treated histone H4 knockdown groups compared to the control group.
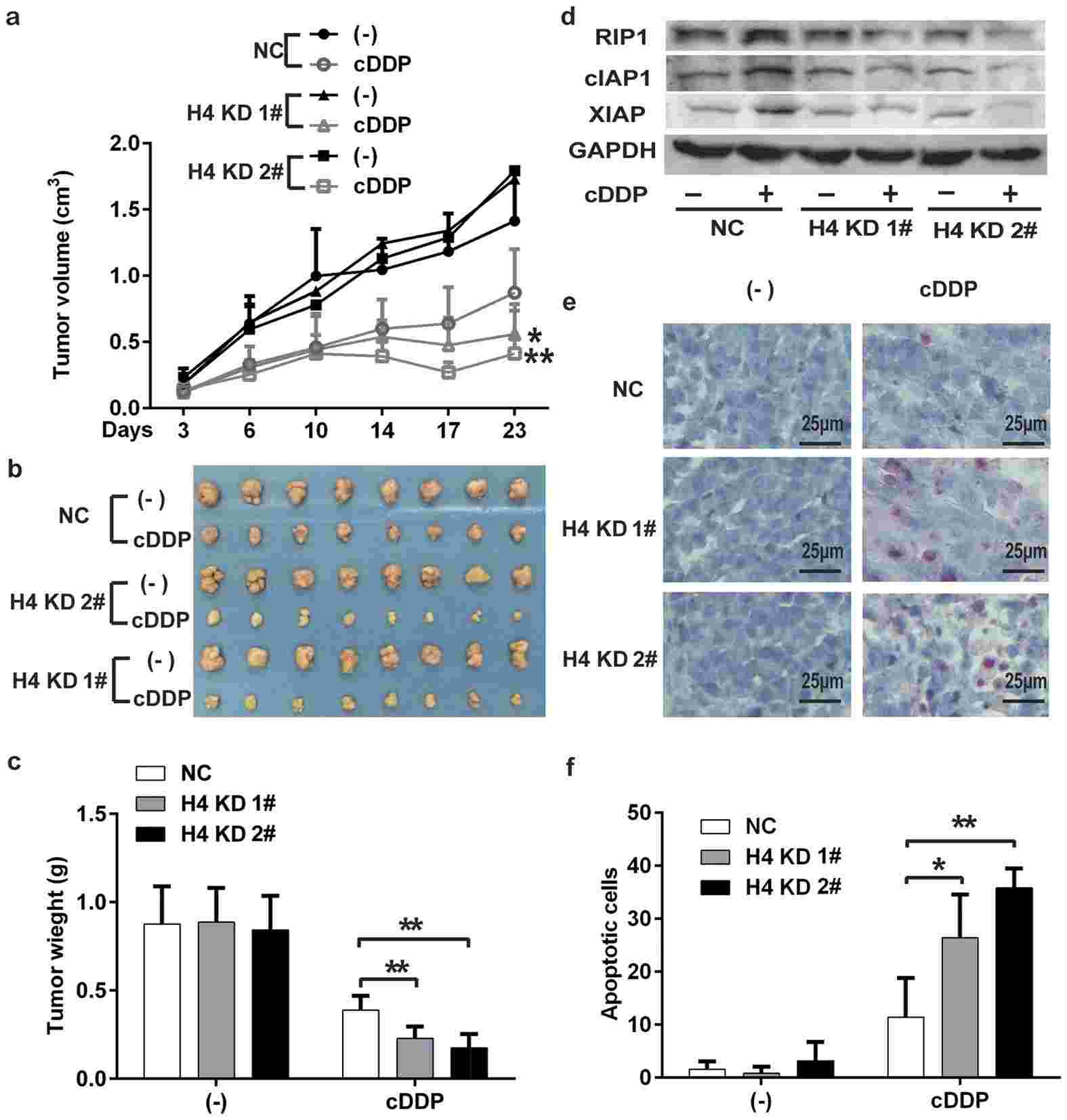 Fig.2 H4 knockdown enhances cisplatin's antitumor activity in vivo.
Fig.2 H4 knockdown enhances cisplatin's antitumor activity in vivo.Related References
- Wakamori M, Fujii Y, Suka N, et al. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep. 2015;5:17204.
- Winogradoff D, Echeverria I, Potoyan DA, Papoian GA. The Acetylation Landscape of the H4 Histone Tail: Disentangling the Interplay between the Specific and Cumulative Effects. J Am Chem Soc. 2015;137(19):6245-6253.
- Garcia BA, Hake SB, Diaz RL, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282(10):7641-7655.

