INO80 Family
Related Symbol Search List
- ACTL6A
- ACTR5
- ACTR6
- Arp8
- BRD8
- DMAP1
- EPC1
- H2AFZ
- HIST2H2BE
- ING3
- INO80
- INO80B
- INO80C
- INO80D
- INO80E
- KAT5
- MCRS1
- MORF4L1
- MORF4L2
- C20orf20
- MRGBP
- NFRKB
- RUVBL1
- RUVBL2
- TFPT
- VPS72
- YEATS4
Immunology Background
Background
The INO80 (Inositol requiring 80) family of chromatin remodelers is a group of ATP-dependent enzymes that play a crucial role in regulating chromatin structure and gene expression. INO80 family is responsible for reorganizing nucleosomes, the structural units of chromatin, to allow for access to DNA by transcription factors and other regulatory proteins.
The INO80 family is named after the INO80 protein, the founding member of the family in yeast. In addition to INO80, the family includes several other proteins, such as SNF2, RUVBL1, and RUVBL2, that work together in complexes to remodel chromatin. These complexes have diverse functions, including regulation of gene expression, DNA repair, and chromosome segregation.
Research has shown that mutations in INO80 family proteins are associated with a variety of human diseases, including cancer, neurodevelopmental disorders, and immune deficiencies. Therefore, understanding the mechanisms of action and regulation of these chromatin remodelers is of great interest for both basic science and potential therapeutic applications.
Structure of INO80 Family Proteins
Structurally, INO80 family members typically consist of multiple subunits that come together to form a large, multi-subunit complex. The core of this complex is formed by the ATPase subunit, which contains the ATPase domain and is responsible for hydrolyzing ATP to drive chromatin remodeling. Surrounding the ATPase subunit are various accessory subunits that help regulate the activity of the complex and facilitate its interactions with chromatin.
Many INO80 family members also contain additional domains that are involved in protein-protein interactions and substrate recognition. For example, some subunits may contain PHD finger domains that recognize specific histone modifications, while others may contain bromodomains that bind to acetylated histones.
The overall structure of INO80 family complexes is highly dynamic and can undergo conformational changes in response to various signals and stimuli. This flexibility allows these complexes to adapt to different chromatin environments and perform their diverse functions.
| Domains | Details |
|---|---|
| Core Components |
INO80 ATPase Subunit: The hallmark of the INO80 family is the presence of the INO80 ATPase subunit, which provides the energy required for chromatin remodeling through ATP hydrolysis. Accessory Subunits: Various accessory subunits surround the ATPase subunit, forming a complex network of interactions that regulate the remodeling activity and specificity of the complex. |
| Subunit Diversity |
Unique Subunit Composition: Different members of the INO80 family can have variable subunit compositions, leading to specialization in distinct cellular functions such as transcription regulation, DNA repair, or replication. Specialized Domains: Some subunits within the complex may harbor specialized domains that facilitate interactions with specific chromatin regions or other protein complexes. |
| Modular Architecture |
Functional Modules: The INO80 family complexes generally exhibit a modular architecture, with distinct subunits forming functional modules responsible for various activities, such as ATP binding, DNA binding, and histone interaction. Regulatory Subunits: Certain subunits may act as regulators, modulating the activity of the core ATPase or influencing the complex's recruitment to specific genomic loci. |
| Interaction Interfaces |
Protein-Protein Interactions: The subunits within the INO80 family complexes interact with each other through specific interfaces, forming a stable and functional chromatin remodeling machinery. Subunit Arrangement: The arrangement of subunits within the complex is critical for proper functioning and coordination of activities like nucleosome sliding, histone exchange, and DNA accessibility modulation. |
| Functional Domains |
ATPase Domain: The INO80 ATPase subunit typically contains ATPase and helicase domains responsible for ATP hydrolysis and DNA translocation activities. Chromatin Binding Domains: Some subunits may contain chromatin-binding domains that enable the complex to interact with nucleosomes and modify chromatin structure. |
| Regulatory Mechanisms |
Post-translational Modifications: Subunits of the INO80 family can undergo post-translational modifications that regulate their activity, stability, or interactions with other proteins. Cofactor Binding Sites: Some subunits may have binding sites for cofactors or nucleic acids, allowing the complex to respond to signals or recruit additional factors for specific functions. |
In summary, the INO80 family of chromatin remodeling complexes exhibits a structured organization with a core ATPase subunit surrounded by accessory subunits that confer functional diversity and specificity. The modular architecture, interaction interfaces, and specialized domains within these complexes play crucial roles in orchestrating chromatin dynamics and gene regulation in various cellular processes.
Role of INO80 Family in Chromatin Remodeling
Here are some key aspects of the role of the INO80 family in chromatin remodeling:
- Nucleosome Sliding: INO80 complexes use energy from ATP hydrolysis to reposition nucleosomes along the DNA strand, allowing access to DNA for various cellular processes such as transcription, replication, and repair.
- Histone Variant Incorporation: INO80 remodelers can replace canonical histones with histone variants, impacting chromatin structure and gene expression regulation.
- DNA Accessibility: By altering nucleosome positioning and stability, INO80 family members facilitate access to DNA sequences for transcription factors, RNA polymerases, and DNA repair machinery.
- Replication Fork Progression: INO80 family members assist in maintaining replication fork stability by evicting nucleosomes ahead of the fork and promoting their reassembly behind it.
- Chromatin Structure Maintenance: INO80 complexes help maintain proper chromatin structure and organization, ensuring genome stability and regulating gene expression patterns.
- Dynamic Chromatin Modifications: INO80 remodelers influence histone modifications and chromatin marks, impacting gene expression profiles and cellular responses to internal and external signals.
In summary, the INO80 family of chromatin remodelers plays a crucial role in dynamically altering chromatin structure to regulate gene expression, DNA repair, replication, and other essential cellular processes. Their activities are vital for maintaining genome integrity, orchestrating proper gene regulation, and ensuring cellular functions operate effectively and accurately.
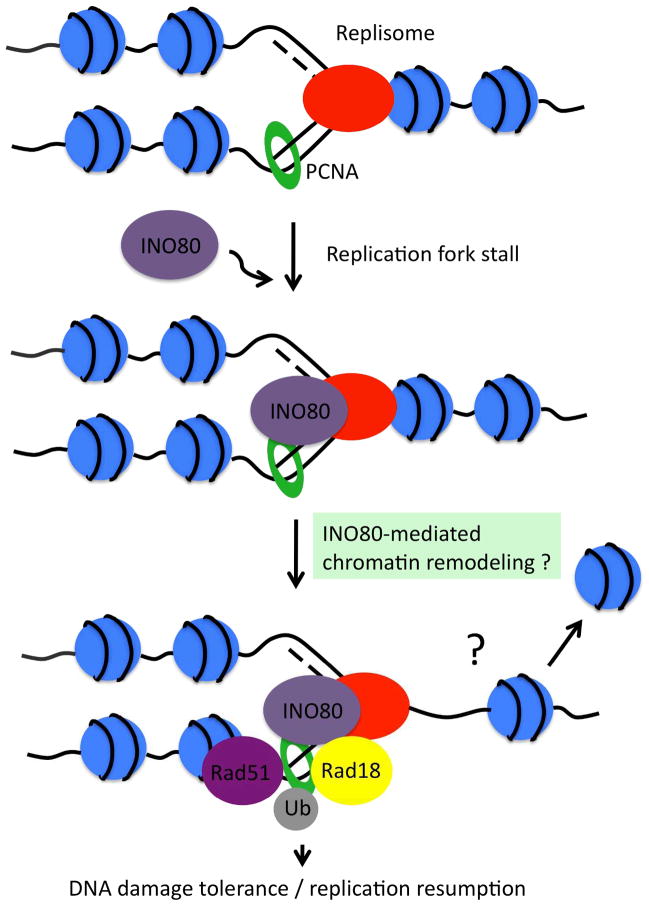 Fig.1 The role of INO80 in DNA damage tolerance during replication. (Falbo KB, et al., 2012)
Fig.1 The role of INO80 in DNA damage tolerance during replication. (Falbo KB, et al., 2012)Functions of INO80 Family Proteins
The INO80 family characterized by their ATP-dependent remodeling activity, plays a crucial role in various physiological processes essential for cell function and genome stability. Here is an overview of their key physiological functions:
| Functions | Details |
|---|---|
| Cell Cycle Regulation |
Chromatin Dynamics: INO80 complexes regulate chromatin structure during the cell cycle, ensuring proper DNA replication, repair, and gene expression. DNA Replication: They facilitate replication fork progression by remodeling chromatin and promoting the assembly of nucleosomes behind the replication machinery. Example: During the G1/S transition, INO80 complexes are involved in preparing chromatin for DNA replication by facilitating nucleosome eviction to allow for DNA polymerases to access the DNA template. |
| DNA Damage Repair |
Homologous Recombination: INO80 family members participate in homologous recombination repair by mobilizing nucleosomes to facilitate access to DNA lesions. Nucleotide Excision Repair: They are involved in nucleotide excision repair pathways, contributing to the removal of bulky DNA lesions. |
| Transcriptional Regulation |
Gene Activation: INO80 complexes aid in gene activation by remodeling chromatin structure at gene promoters, enhancers, and other regulatory regions. Gene Repression: They also play a role in gene repression by modulating chromatin accessibility and nucleosome positioning. |
| Epigenetic Regulation |
Histone Modifications: INO80 remodelers influence histone modifications that regulate gene expression patterns and chromatin structure. Chromatin Accessibility: They modulate chromatin accessibility to transcription factors and regulatory proteins, impacting gene transcription. |
| Cell Differentiation and Development |
Embryonic Development: INO80 complexes are involved in embryonic development by controlling gene expression programs critical for cell differentiation and tissue development. Stem Cell Maintenance: They contribute to maintaining stem cell pluripotency and self-renewal by regulating chromatin dynamics. |
| Genome Stability |
Maintaining Chromatin Integrity: INO80 family members ensure proper chromatin organization and stability, which is crucial for preserving genomic integrity. Preventing Transposon Activation: They help suppress transposable elements within the genome, preventing genomic instability and maintaining genome integrity. |
| Response to Environmental Stress | Stress Response: INO80 complexes are involved in the cellular response to environmental stresses by modulating chromatin structure and gene expression to adapt to changing conditions. |
| Regulation of Non-Coding DNA |
Enhancer Regulation: INO80 remodelers participate in the regulation of enhancer activity by modulating chromatin structure and accessibility at enhancer regions. Long Non-Coding RNA Regulation: They influence the expression of long non-coding RNAs by modulating chromatin architecture at non-coding regions of the genome. |
| Disease Implications |
Cancer: Dysregulation of INO80 family members is associated with cancer development and progression due to their roles in transcriptional regulation and genome stability. Neurodevelopmental Disorders: Mutations in INO80 complexes have been linked to neurodevelopmental disorders, highlighting their importance in brain development and function. |
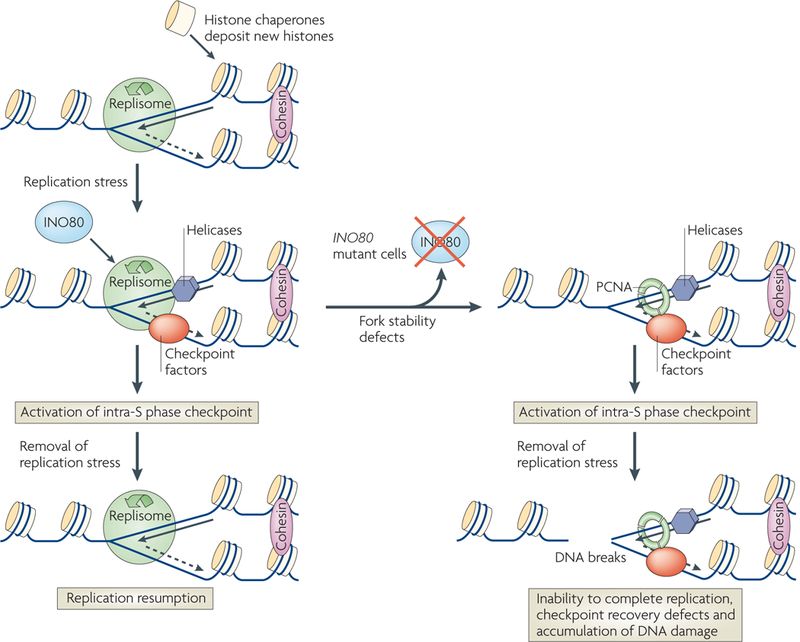 Fig.2 The INO80 complex promotes recovery of stalled replication forks. (Morrison AJ, et al., 2009)
Fig.2 The INO80 complex promotes recovery of stalled replication forks. (Morrison AJ, et al., 2009)Members and Features of INO80 Family
Here are some of the main INO80 family type members and their features:
| Types | Members and features |
|---|---|
| Core INO80 Complex Members |
|
| Other INO80 Family Members |
|
| Other Related Molecules |
|
These are just some examples of the members of the INO80 family that, together, form the INO80 family and are involved in regulating chromatin structure and function, as well as regulating gene expression and cellular processes. Each member plays a specific role in the complex, working together in harmony to maintain genome stability and proper function within the cell.
INO80 Family in the Occurrence and Development of Diseases
Dysregulation of INO80 family members has been linked to various diseases, including cancer, neurological disorders, and developmental defects. Here are some examples of how the INO80 family is involved in the occurrence and development of diseases:
Cancer
INO80 has been shown to be upregulated in various types of cancer, including prostate, breast, and liver cancer. INO80 promotes tumor growth and metastasis by regulating the expression of genes involved in cell proliferation and invasion. For example, in breast cancer, INO80 promotes the expression of genes involved in cell cycle progression and DNA repair, leading to increased tumor growth and resistance to chemotherapy.
Neurological Disorders
Mutations in INO80 family genes have been linked to neurodevelopmental disorders such as Coffin-Siris syndrome, a rare genetic disorder characterized by intellectual disability, developmental delay, and distinct facial features. INO80 complexes have been shown to be important for neuronal development and function, and disruptions in INO80 activity can lead to abnormal brain development and neurological dysfunction.
Developmental Defects
INO80 complexes are essential for normal embryonic development and cell differentiation. Mutations in INO80 family genes can result in developmental defects such as craniofacial abnormalities, heart defects, and other birth defects. For example, ino80 knockout mice exhibit severe developmental defects and embryonic lethality, highlighting the importance of INO80 in embryonic development.
Overall, the INO80 family of chromatin remodeling complexes plays a crucial role in various physiological processes, and dysregulation of INO80 activity can contribute to the occurrence and development of diseases. Further investigation of the molecular mechanism of INO80 function may provide new therapeutic strategies for the treatment of INO80-related diseases.
Case Study
Case 1: Thang NX, Han DW, Park C, et al. INO80 function is required for mouse mammary gland development, but mutation alone may be insufficient for breast cancer. Front Cell Dev Biol. 2023;11:1253274.
The aberrant function of the ATP-dependent chromatin remodeler INO80 has been linked to various types of cancers through the alteration of chromatin structure and gene expression. Nevertheless, the precise mechanism by which mutations in INO80 contribute to cancer development, particularly in breast cancer, remains unclear.
In the current investigation, a weighted gene co-expression network analysis (WGCNA) was conducted to explore the relationship between the expression of INO80 and the subtypes and progression of breast cancer. The analysis demonstrated that the repression of INO80 is connected to the varying response of estrogen receptors (ERs) based on the subtype of breast cancer, ER networks, and an increased susceptibility to breast cancer development.
To assess whether the loss of INO80 triggers the formation of breast tumors, researchers generated a conditional INO80-knockout (INO80 cKO) mouse model utilizing the Cre-loxP system. Phenotypic evaluation of the model revealed that INO80 cKO resulted in reduced branching and length of mammary ducts across all developmental stages. However, the INO80 cKO mouse model did not display any changes in the morphology of the mammary gland's lumens and did not spontaneously induce tumorigenesis.
Conclusively, the study suggests that the aberrant function of INO80 may be linked to breast cancer by influencing gene expression. However, it was observed that INO80 mutation alone is inadequate to induce breast tumorigenesis.
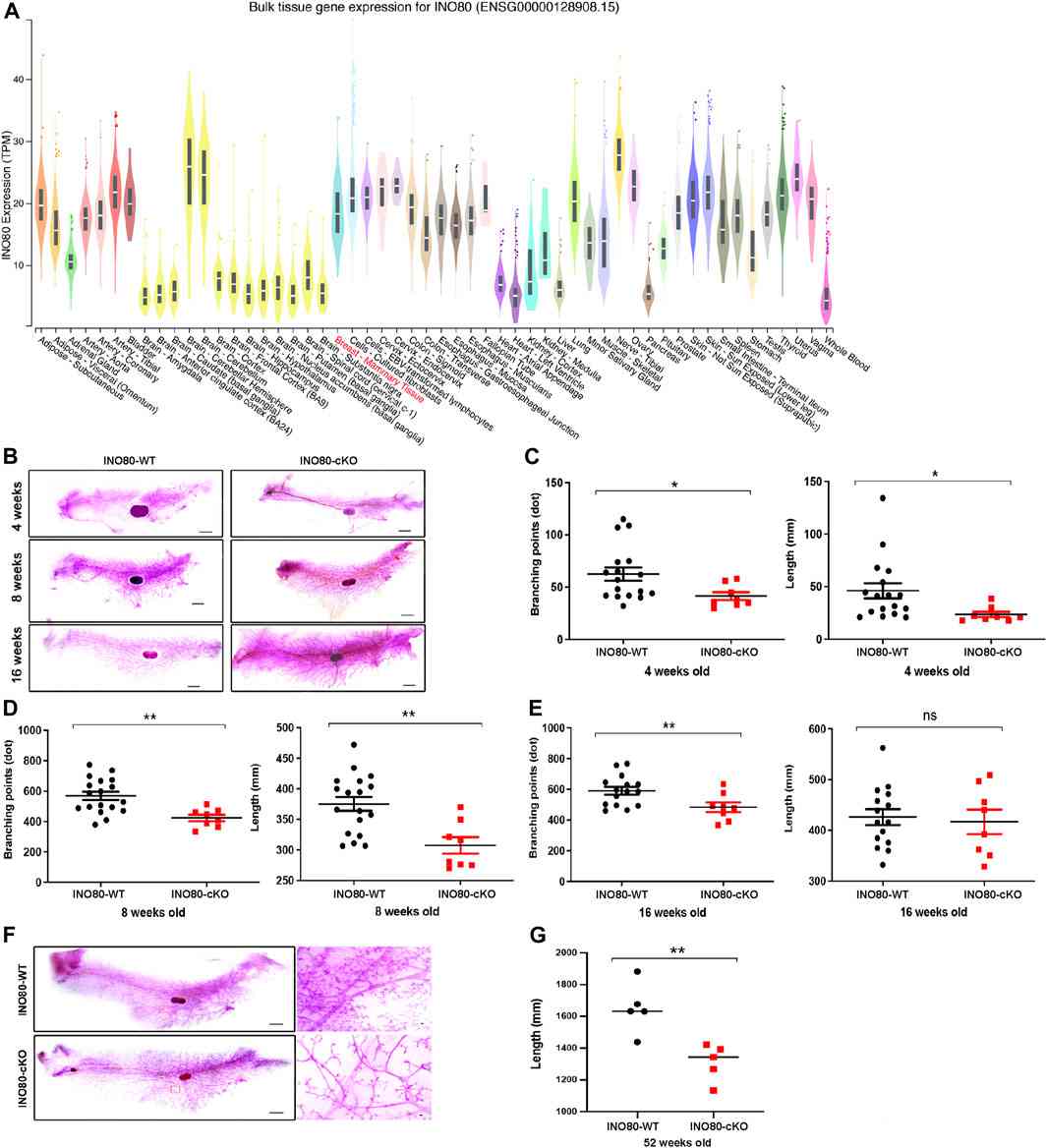 Fig.1 INO80 loss impairs mouse mammary gland development.
Fig.1 INO80 loss impairs mouse mammary gland development.Case 2: Yoo S, Lee EJ, Thang NX, La H, Lee H, Park C, Han DW, Uhm SJ, Song H, Do JT, et al. INO80 is required for the cell cycle control, survival, and differentiation of mouse ESCs by transcriptional regulation. International Journal of Molecular Sciences. 2022; 23(23):15402.
Precise regulation of the cell cycle of embryonic stem cells (ESCs) is crucial for their self-maintenance and differentiation. The cell cycle of ESCs differs from that of somatic cells and varies depending on the cell culture conditions. However, the mechanisms by which epigenetics regulates the cell cycle in ESCs are not fully understood. In a recent study, researchers have demonstrated that the ATP-dependent chromatin remodeler Ino80 plays a key role in regulating cell cycle genes in ESCs under primed conditions.
Loss of Ino80 results in a significant extension of the G1-phase in ESCs grown under primed culture conditions. Ino80 directly binds to the transcription start site of cell cycle-related genes and controls their expression, leading to cell cycle progression. Additionally, loss of Ino80 triggers cell apoptosis in ESCs. Interestingly, the role of Ino80 in regulating the cell cycle during ESC differentiation is slightly different, as inducible knockout of Ino80 results in an extended S-phase in differentiating ESCs.
Further analysis using RNA-seq revealed that the expression of genes involved in organ development and cell cycle regulation is persistently altered in Ino80 knockout cells during differentiation, indicating that Ino80's role in cell cycle regulation is not limited to undifferentiated ESCs. This study highlights the importance of Ino80 in the regulation of the ESC cycle through transcriptional mechanisms, which may also be applicable to other cell types.
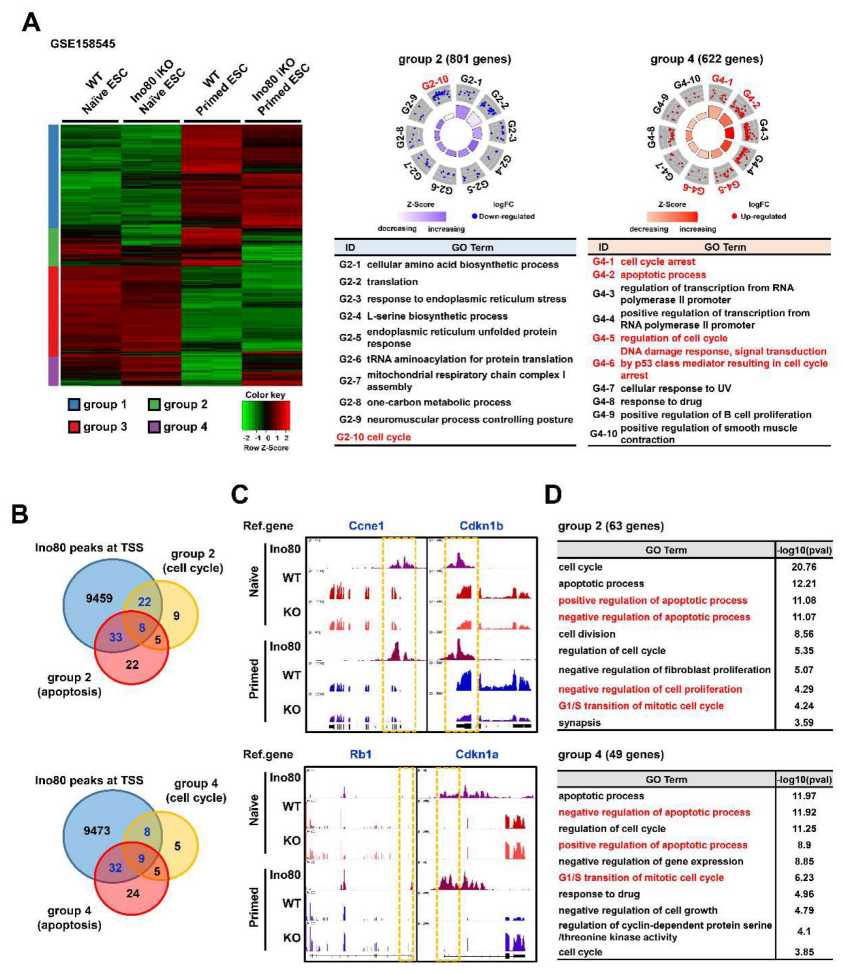 Fig.2 Ino80 directly regulates genes involved in the cell cycle.
Fig.2 Ino80 directly regulates genes involved in the cell cycle.Related References
- Watanabe S, Peterson CL. The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb Symp Quant Biol. 2010;75:35-42.
- Falbo KB, Shen X. Function of the INO80 chromatin remodeling complex in DNA replication. Front Biosci (Landmark Ed). 2012;17(3):970-975.
- Morrison AJ, Shen X. Chromatin remodeling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10(6):373-384.
- Yoo S, Lee EJ, Thang NX, La H, Lee H, Park C, Han DW, Uhm SJ, Song H, Do JT, et al. INO80 is required for the cell cycle control, survival, and differentiation of mouse ESCs by transcriptional regulation. International Journal of Molecular Sciences. 2022; 23(23):15402.

