Principle and Protocol of Yeast Two-hybrid System
The classical yeast two-hybrid system requires that the measured proteins be located in the nucleus. However, some proteins have strong hydrophobic domains, and some proteins carry strong signals located in other organelles of the cell, so they cannot be transported to the nucleus. The interaction between proteins cannot be detected by this system; Secondly, the system detects the interaction between proteins by activating the transcription reporter gene. If the protein is a regulatory transcriptional protein (such as a suppressor) that inhibits gene expression, the reporter gene cannot be activated for transcription, and the interaction between the protein and other proteins cannot be detected by the two-hybrid system. At the same time, this system cannot detect the protein-protein interaction and the interaction between cytoplasmic and membrane proteins that require post-transcriptional translation in the endoplasmic reticulum or cytoplasm. Therefore, this system has certain limitations.
In recent years, many new yeast two-hybrid systems have been derived from the classical yeast two-hybrid system, such as the Sos recruitment system. Unlike the traditional yeast two-hybrid system, the Sos recruitment system needs to rely on transcriptional activation to detect the interaction between proteins. This system can detect the interaction between transcriptional factor activators or inhibitors. In addition, the protein-protein interaction detected by the Sos recruitment system occurs in the thorax. First, it can be used to detect the interaction between some proteins that cannot be well located in the nucleus. Second, it can allow the detection of post-transcriptional modified proteins. For some proteins, this is necessary for the system to truly reflect the protein-protein interaction. After Aronheim applied the Sos recruitment system to screen the protein JDP2 that interacts with the transcription factor AP-1, more and more studies have used this system for the discovery of new genes. For example, Ko et al. used the binding domain of thyroxine receptor β1 ligand as a "bait" protein and screened a protein TRBP binding to thyroxine receptor. The system has also been well applied in the research of plant flower development, disease prevention and ion channels.
The experimental manual mainly introduces the application of the Sos recruitment system to screen interacting proteins.
The purpose of this experiment manual is to help scientific researchers master the principle and application of the yeast two-hybrid Sos recruited system, learn to use the co-transformation method to screen the interacting proteins, and be familiar with the operation process and precautions of the yeast two-hybrid system for library screening.
So recruitment system is a new yeast two-hybrid system. This system is a two-hybrid system designed by Aronheim and others according to the characteristics of Sos protein. Sos protein is the guanine nucleoside exchange factor (GEF) in human Ras pathway, which is located in the cytoplasm. During the process of cell signal transduction, it can convert Ras protein from inactive Ras-GDP to activated Ras-GTP, thus activating Ras signal pathway and enabling cell growth. GEF in yeast is Cde25 protein, which is homologous with human Sos protein and has the same function as Sos protein. Its mutant Cdc25 is temperature-sensitive and cannot grow at 37°C. It was found that in mutant Cdc25 yeast cells, when Sos protein was located on the cell membrane in a certain way, it could replace the function of Cdc25 protein, activate Ras protein to restore cell growth at 37°C. In this system, Sos protein and target protein X are fused and expressed, and decoy protein Y is fused and expressed with myristylation signal (Myr), so that Y is located on the membrane. If there is an interaction between X and Y, the Sos protein is recruited to the cell membrane, thus activating the Ras signal pathway and making yeast cells grow at 37°C. On the contrary, no yeast colony can be observed (Figure 4-2-1).
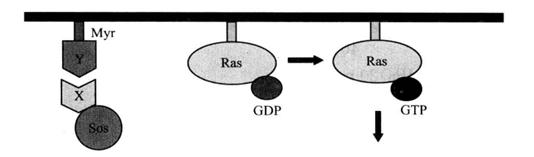 Figure 4-2-1 Schematic Diagram of Yeast Two-Hybrid Sos Recruitment System Principle
Figure 4-2-1 Schematic Diagram of Yeast Two-Hybrid Sos Recruitment System Principle
1. Main Instruments and Equipment
Micropipette (2 μL, 10 μL, 20 μL, 200 μL, 1000μL), low-temperature centrifuge, desktop centrifuge, agarose gel electrophoresis system, protein gel electrophoresis system, gel imaging system, semi-dry transfer system, thermostatic shaker, thermostatic incubator, fume hood, ice maker, oscillator, thermostatic metal bath, sterile inoculation ring, 10 cm culture dish, 15 cm culture dish.
2. Experimental Materials
Yeast strain*1, yeast expression vector*2.
3. Main Reagents
(1) YPDA liquid culture medium: YPD medium (0.05 kg/L), adenine sulfate (0.04 g/L), high-pressure steam sterilization (121°C, 0.105MPa) for 15min.
(2) YPDA solid medium: YPD agar medium (0.07 kg/L), adenine sulfate (0.04 g/L), high-pressure steam sterilization (121°C, 0.105MPa) for 15min.
(3) SD/glucose (-UL) solid medium: minimal SD agar base (46.7 g/L), -His/-Leu/-Ura DO supplement (0.65 g/L), histidine (0.2g/L), high-pressure steam sterilization (121°C, 0.105MPa) for 15min.
(4) SD/galactose (-UL) solid medium: minimal SD agar base/Gal/Ra (54 g/L), -His/-Leu/-Ura DO supplement (0.65 g/L), histidine (0.2 g/L), high-pressure steam sterilization (121°C, 0.105MPa) for 15min.
(5) 1mol/L sorbitol: weigh 18.2g of sorbitol and dissolve in 100mL of deionized water, high pressure steam sterilization (121°C, 0.105MPa) for 20min.
(6) Adenine, PEG 3350, vector DNA, DMSO.
(7) LiSORB solution
| 1 mol/L | Sorbitol |
| 100 mmol/L | LiAc |
| 10 mmol/L | Tris-HCl (pH 8.0) |
| 1 mmol/L | EDTA |
| High-pressure steam sterilization (121°C, 0.105MPa) for 20min. | |
(8) Z-buffer / X-β-gal solution
| 10 mmol/L | Tris-HCl (pH 8.0) |
| 100 mmol/L | LiAc |
| 1 mmol/L | EDTA |
| 40% | PEG 3350 |
| High-pressure steam sterilization (121°C, 0.105MPa) for 20min. | |
1. Yeast recovery
(1) 1-2 days before resuscitation, prepare YPDA agar and reverse the plate after high-pressure disinfection.
(2) Select a small group of yeast cells from the yeast cryopreservation tube with sterile contact ring, and activate cdc25H (α) strain by lineation.
(3) Incubate at room temperature (22-25°C) upside down for 4-6 days.
2. Preparation of cdc25H yeast receptive cells
(1) Select a yeast clone with a diameter of 2-3mm and inoculate it into 1mL YPDA liquid medium.
(2) The violent oscillation makes the cell mass disperse evenly.

(3) Transfer the yeast cell suspension into a conical flask containing 50mL of fresh YPDA liquid medium, and culture at room temperature at 230r/min for 16h until the stable period (A600>1).
(4) Transfer the appropriate overnight culture into a conical flask containing fresh 300mL YPDA medium to make its A600 about 0.2.
(5) At room temperature, 250 r/min, shake and incubate to make its A600 greater than 0.7.
(6) Take 75 μL culture was coated on YPAD plate and cultured at 37°C*3. Transfer the remaining yeast culture into several 50mL centrifuge tubes at room temperature, 1000g, and centrifuge for 5min.
(7) Room temperature, 1000g, centrifugation for 10 min.
(8) Discard the supernatant, resuspend cells with 50mL LiSORB, and place them at room temperature for 30min.
(9) Within 30 min of cultivating yeast cells, 480 μL10mg/ml salmon sperm DNA was treated in boiling water bath for 20 minutes. Add 120 μL LiSORB, suck, and beat well. Cool to room temperature.
(10) After 30min at room temperature, 1000g at room temperature, and centrifugation for 5min.
(11) Use 300 μL LiSORB resuspended yeast cells.
(12) Join 600 μL the salmon sperm DNA mixture prepared in step (10) is gently beaten and thoroughly mixed.
(13) Add 5.4mL PEG/LiOAc solution and 530 μL DMSO. Gently suck and beat, thoroughly mix, and reserve *4.
3. Screening of "bait" protein library
(1) Divide 5mL of freshly prepared yeast receptive cells into a 50mL tube, and add 20μg of pSos-bait "bait" protein particles*5, 20μg pMyr-cDNA library plasmid and 100 μL of 1.4mol/L β-mercaptoethanol, gently invert or knock, and thoroughly mix.
(2) Dispense into 10 1.5mL tubes of 500μL each.
(3) Leave at room temperature for 30min, mixing gently several times in between.
(4) Heat shock at 42°C for 20min.
(5) Place on ice for 3 min.
(6) Centrifuge at room temperature, 14000 r/min for 30s and collect the cells.
(7) Discard the supernatant and add 500 μL of 1 mol/L sorbitol to resuspend the cells.
(8) Apply on 150mm SD/glucose (-UL) plates and incubate at room temperature (22-25°C) for 48h.
(9) Yeast cells were copied onto SD/galactose (-UL) plates with sterilized filter paper and incubated at 37°C for 6-10 d.
(10) Pick yeast clones from SD/galactose (-UL) plates, resuspend in 25 μL of sterilized water, take 2.5 μL and add dropwise to SD/glucose (-UL) plates and incubate at room temperature (22-25°C) for 48h.
(11) Yeast clones were picked, resuspended in 25 μL sterilized water, and 2.5 μL was added dropwise to 2 SD/glucose (-UL) plates and one SD/galactose (-UL) plate, respectively.
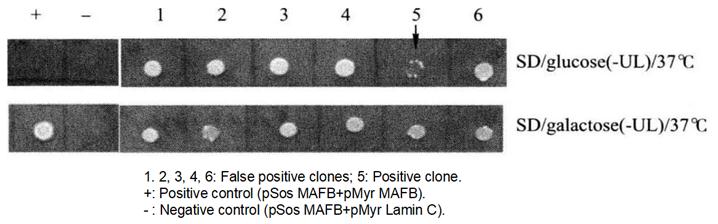 Figure 4-2-2 Screening of Positive Yeast Clones
Figure 4-2-2 Screening of Positive Yeast Clones
(12) One SD/glucose (- UL) plate and SD/galactose (- UL) plate were cultured at 37°C for 4-6 days as the first round of validation.
(13) Another SD/glucose (- UL) plate was placed at room temperature (22-25°C) for 48h for the second round of validation.
(14) Clones were picked from another SD/glucose (-UL) plate, resuspended in 25 μL sterilized water, and 2.5 μL were added dropwise to SD/glucose (-UL) and SD/galactose (-UL) plates, respectively, and incubated at 37°C for 4-d. This was the second round of validation.
4. Positive clone verification
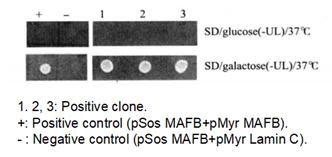 Figure 4-2-3 Verification of Positive Yeast Clones
Figure 4-2-3 Verification of Positive Yeast Clones
(1) Pick a single positive clone in 5mL SD/glucose (-UL) liquid medium and incubate at room temperature (22-25°C) with shaking at 250r/min for 2-3d.
(2) Extract yeast plasmids*6.
(3) The extracted plasmid mixture (BD/bait, AD/cDNA) was transformed into Top10 E. coli using the classical transformation method by spreading LB antibiotic plates. the antibiotics in the LB plates were consistent with the resistance carried by the pMyr/cDNA vector, so the final clone obtained contained only pMyr/cDNA.
(4) Pick a single positive clone in 5mL LB medium (with corresponding antibiotics) and incubate at 37°C, 250-270r/min, with shaking for 12-16h.
(5) Extract the plasmid and do PCR for identification.
(6) Co-transform the pSos-bait expression plasmid and the pMyr-cDNA expression plasmid into Cdc25H(α) yeast to verify the authenticity of the positive clones*7.
*1 Different yeast two-hybrid systems with different yeast strains available. Here we choose cdc25H (α) and cdc25H (A) yeast strains.
*2 The yeast expression plasmid used in this system is from the CytoTrap system of Stratagene Company.
*3 Yeast cells were cultured at 37°C for 4 to 6 days, and temperature-sensitive reversion mutations were detected.
*4 Yeast-receptive cells should be used immediately and should not be stored at - 80°C for a long time.
*5 The "bait" protein particles should first pass the self-activation test and then be screened for the library.
*6 Use commercial yeast extraction kit to extract yeast plasmid.
*7 Co-conversion into yeast, temperature screening, and detection of protein-protein interaction.

