Codon Optimization
Background
Efficient gene expression is a cornerstone of successful recombinant protein production. However, discrepancies between native gene sequences and the preferences of a heterologous host often lead to suboptimal protein yield. Codon usage bias, mRNA stability, and regulatory elements all contribute to this expression variability.
To address these challenges, codon optimization is employed as a powerful strategy to redesign gene sequences for enhanced expression efficiency—without altering the encoded protein’s amino acid sequence. This approach is essential for improving yields in research, diagnostics, therapeutics, and industrial biotechnology.
Creative BioMart offers a cutting-edge Codon Optimization Service using our proprietary gene optimization technology, developed through years of innovation and expertise in protein expression. Our system is engineered to enhance the expression of both naturally occurring and recombinant gene sequences across various host systems.
The service is tailored for clients seeking improved productivity in bacterial, yeast, insect, mammalian, or other custom expression platforms. By factoring in critical biological and computational parameters, we ensure a significant boost in both transcriptional and translational efficiency of target genes.
What We Offer?
-
Key Features
- Codon Usage Adaptation
- mRNA Structure Optimization
- Transcriptional & Translational Element Adjustments
- Elimination of Unwanted Motifs
-

Deliverables
- Optimized nucleotide sequence (DNA)
- Detailed optimization report with comparison to native sequence
- Optional: Synthetic gene synthesis and subcloning into your vector of choice
- Optional: Expression testing and protein production in your selected host
-
Supported Hosts
- Escherichia coli
- Saccharomyces cerevisiae / Pichia pastoris
- Insect cells (Sf9, Sf21)
- Mammalian cells (CHO, HEK293)
- Others upon request
-

Turnaround Time
- Codon optimization only: 3–5 business days
- With synthesis and subcloning: 2–3 weeks
Why Choose Us?
- Proprietary Algorithm: Scientifically validated optimization system refined by years of research.
- Expertise in Expression: Deep knowledge of host-specific gene expression mechanisms.
- Proven Results: Consistent improvement in expression yields across diverse systems.
- Full-Service Capability: From sequence design to gene synthesis and expression validation.
- Confidential & Reliable: Trusted by global biotech and pharmaceutical partners.
- Customized Solutions: Tailored optimization strategies to meet unique project goals and host system requirements.
Case Study
* NOTE: We prioritize confidentiality to safeguard our clients’ technology and intellectual property. As an alternative, we present selected published research articles as representative case studies. For details on the assay services and products used in these studies, please refer to the relevant sections of the cited literature.
Case 1: Codon pair optimization (CPO) for protein expression in Pichia pastoris
Huang et al., 2021. doi:10.1186/s12934-021-01696-y
This study highlights the importance of codon-pair context in optimizing protein expression in Pichia pastoris. While traditional codon optimization focuses on replacing rare codons with frequently used ones, this research demonstrates that codon-pair bias—which influences translation efficiency and accuracy—has a greater impact.
Using a novel Codon Pair Optimization (CPO) algorithm, researchers designed and tested multiple gene variants for two scFv proteins. Variants optimized for favorable codon-pair context showed 5- to 7-fold higher expression than those optimized by standard codon usage. These findings support codon-pair-based gene design as a superior strategy for enhancing recombinant protein yield in P. pastoris.
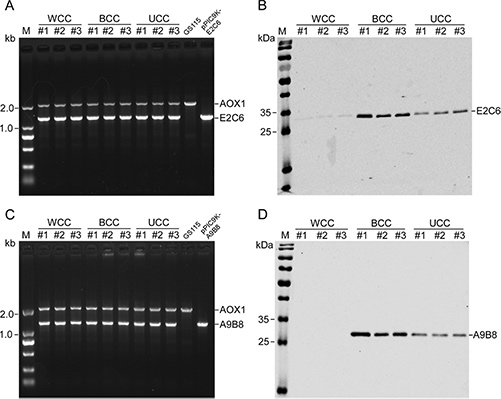
Figure 1. The expression of variants of MT1-MMP E2C6 and ADAM17 A9B8 with the worst, the best, and the unbiased codon-pair context in Pichia pastoris. A, C Analysis of integrants using AOX1 general primers by PCR. Lane GS115 is a control using the host GS115 genome as the template, which show AOX1 gene, and lane pPIC9K-E2C6 or pPIC9K-A9B8 is another control using recombinant plasmid pPIC9K-E2C6 or pPIC9K-A9B8 as the template, which shows synthetic gene of MT1-MMP E2C6 and ADAM17 A9B8. B, D The expression of variants in Pichia pastoris. MT1-MMP E2C6 and ADAM17 A9B8 in the supernatant are detected by Western blot using anti-his tag primary antibody. (Huang et al., 2021)
Case 2: Efficient retroviral pseudotyping with codon-optimized chimeric GALV envelope
Mirow et al., 2021. doi:10.3390/v13081471
The Gibbon Ape Leukemia Virus envelope protein (GALV-Env) is a valuable tool for gene therapy due to its ability to transduce human B and T cells. Researchers developed a codon-optimized variant of a chimeric GALV-Env (coGALV-Env) to improve expression and vector titers. This optimized version enabled efficient pseudotyping of γ-retroviral, lentiviral, and α-retroviral vectors with significantly lower plasmid input—only 20 ng per million HEK293T cells.
Importantly, coGALV-Env vectors effectively transduced primary human T cells, making it ideal for applications like CAR-T or TCR engineering. Its efficiency and lower plasmid demand offer a cost-effective solution for GMP-grade vector production.
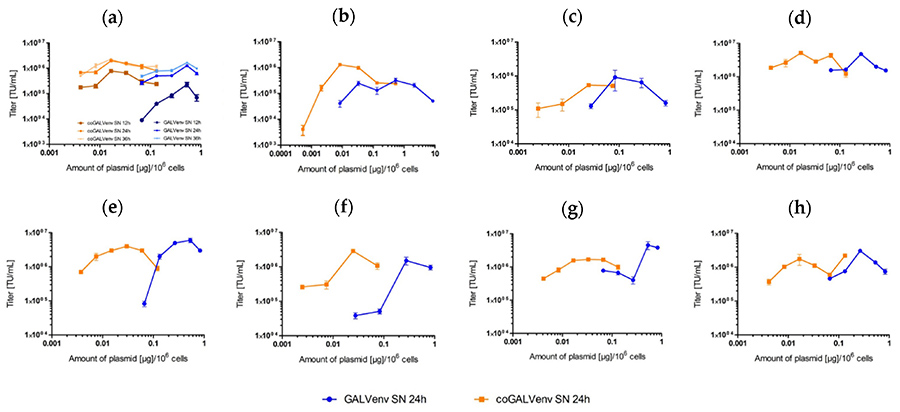
Figure 2. Comparative titration of lentiviral LeGO-mOrange2 vectors produced with standard vs. codon-optimized (co) GALV-env. (a) Much lower amounts of coGALV-env as compared to GALV-env were required to obtain maximal titers. Supernatants were harvested 12, 24, and 36 h after transfection of 293T cells. As evident, maximal titers were in all cases obtained with >20 times lower amounts of coGALV-env as compared to GALV-env; (b–h) titration experiments for seven additional, independent vector productions (only the 24 h data point is shown). Means and standard deviations of triplicates are shown. Small error bars are part hidden behind data point symbols. (Mirow et al., 2021)
Customer Testimonials
-
"Creative BioMart significantly improved the expression of a GC-rich gene we were struggling with in E. coli. Their codon optimization not only replaced rare arginine codons but also adjusted the GC content for better transcriptional efficiency. Expression levels tripled."
— Senior Scientist | Biotech Startup Focused on Protein Therapeutics
-
"We needed codon optimization for a mammalian expression system to produce a viral envelope protein for vaccine development. Creative BioMart delivered a redesigned gene that improved protein folding and secretion in HEK293 cells. Turnaround time and support were excellent."
— Director of Vaccine R&D | Government Research Institute
-
"Our team was producing antibody fragments in Pichia pastoris, but yields were inconsistent. Creative BioMart’s codon optimization aligned the gene with yeast codon usage bias and removed inhibitory sequence motifs. Final yield increased 4-fold with minimal project delays."
— Principal Investigator | University Lab Specializing in Synthetic Biology
-
"We had a tough gene with multiple internal TGA codons that limited translation in our insect cell line. Creative BioMart’s optimization corrected these without altering protein function, and expression in Sf9 cells was robust and reproducible."
— Head of Molecular Biology | Mid-Sized Pharma Company
-
"Codon optimization for a multi-epitope fusion protein in CHO cells was critical for our preclinical pipeline. Creative BioMart tailored the sequence, balanced GC content, and ensured optimal Kozak context. The result was a tenfold increase in expression and cleaner purification."
— VP of Biologics Development | Global Contract Research Organization (CRO)
FAQs
-
Q: Will codon optimization change the protein sequence?
A: No. Codon optimization alters only the nucleotide sequence while preserving the original amino acid sequence of the protein. -
Q: Can you work with sequences from rare or poorly characterized organisms?
A: Yes. Our optimization platform accommodates diverse organisms and expression hosts. Custom evaluations are available. -
Q: Do you offer gene synthesis and expression testing as part of the package?
A: Absolutely. We provide optional gene synthesis, vector construction, and expression validation services upon request. -
Q: How do I submit a sequence for optimization?
A: You can upload your gene sequence in FASTA format through our secure online portal or contact our technical team for assistance. -
Q: What information do you need to start the optimization process?
A: Simply provide the DNA or protein sequence and the intended expression host. We’ll handle the rest.
Resources
Related Services
- Protein Expression and Purification Services
- Mammalian Expression Systems
- Yeast Expression Systems
- Baculovirus-Insect Cell Expression
- Bacterial Expression Systems (E. coli / Bacillus)
- Serum-Free Expression System
- Cell-Free In Vitro Protein Expression
- Fc Fusion Protein Production
- Protein Engineering (Mutant Libraries)
- Sequence Based Protein Design
Related Products
References:
- Huang Y, Lin T, Lu L, et al. Codon pair optimization (CPO): a software tool for synthetic gene design based on codon pair bias to improve the expression of recombinant proteins in Pichia pastoris. Microb Cell Fact. 2021;20(1):209. doi:10.1186/s12934-021-01696-y
- Mirow M, Schwarze LI, Fehse B, Riecken K. Efficient pseudotyping of different retroviral vectors using a novel, codon-optimized gene for chimeric GALV envelope. Viruses. 2021;13(8):1471. doi:10.3390/v13081471
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

