Chip-Tip Technology - Revolutionary Progress in Breaking Through the Bottleneck of Single Cell Proteomics Research
Chip-Tip, a breakthrough in single-cell proteomics (SCP) that dramatically enhances sensitivity and scalability. The method enables the identification of over 5,000 proteins in individual HeLa cells and up to 6,500 proteins when combined with high-field asymmetric waveform ion mobility spectrometry (FAIMS). Notably, it facilitates the direct detection of post-translational modifications (PTMs) like phosphorylation and glycosylation without prior enrichment—a first for SCP. The workflow integrates the cellenONE platform for single-cell isolation, the proteoCHIP EVO 96 for low-loss sample preparation, the Evosep One LC system for rapid chromatography, and the Orbitrap Astral mass spectrometer with narrow-window data-independent acquisition (nDIA) for high-sensitivity analysis. By optimizing parameters such as isolation windows and ion injection times, the authors achieved unprecedented proteome depth. Applications include studying drug-treated cancer spheroids (e.g., 5-fluorouracil effects) and human-induced pluripotent stem cell (hiPSC) differentiation, revealing lineage-specific markers and dynamic proteomic changes. The workflow's throughput reaches 120 samples per day, setting a new benchmark for SCP scalability.
Single-cell proteomics (SCP) has emerged as a transformative tool for dissecting cellular heterogeneity, yet technical limitations in sensitivity and throughput have hindered its broad adoption. The Chip-Tip workflow, detailed in this study, addresses these challenges through a suite of innovations that redefine the boundaries of SCP.
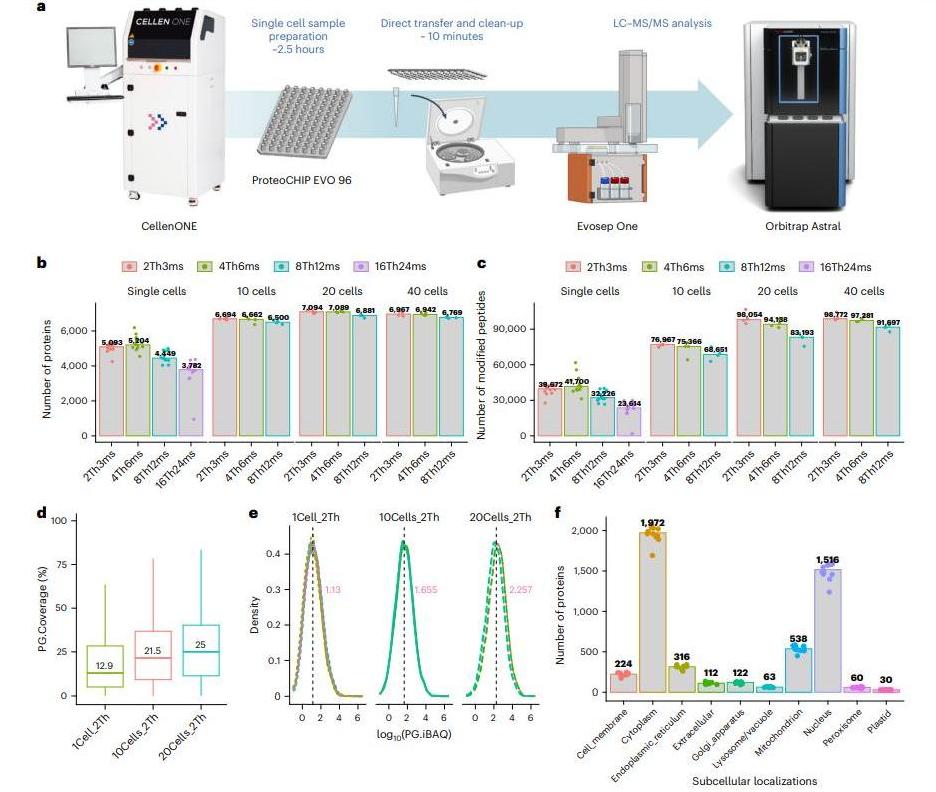
The Chip-Tip method combines precision instrumentation and computational strategies to minimize sample loss and maximize detection. Key advancements include:
- Single-Cell Handling: The cellenONE platform isolates cells with high accuracy, while the proteoCHIP EVO 96 enables nanoliter-scale sample preparation, reducing protein adsorption and evaporation.
- Enhanced Mass Spectrometry: Coupling the Evosep One LC system with the Orbitrap Astral mass spectrometer and nDIA improves chromatographic resolution and sensitivity. Narrow isolation windows (4 Th) and optimized ion injection times yield deep proteome coverage—41,700 peptides per single cell—with a dynamic range spanning four orders of magnitude.
- Carrier Proteome Strategy: Incorporating data from bulk samples (e.g., 20-cell aggregates) boosts single-cell identifications by 20–30%, though this introduces trade-offs in quantification consistency between tools like Spectronaut and DIA-NN.
The workflow's utility is demonstrated in two contexts:
- Drug Response in Spheroids: Treatment with 5-fluorouracil (5-FU) disrupted cancer spheroid integrity, with proteomic profiling revealing upregulated enzymes (e.g., TYMP) linked to drug activation and downregulated structural proteins (e.g., keratins).
- hiPSC Differentiation: Chip-Tip quantified low-abundance transcription factors (OCT4, SOX2) in hiPSCs and lineage-specific markers (GATA4, HAND1, MAP2) in differentiating embryoid bodies. Clustering analysis uncovered pathways tied to pluripotency loss, such as mitochondrial respiration and chromatin remodeling.
While the study marks a leap forward, limitations remain. Throughput is constrained by MS acquisition rates, and PTM identification—though groundbreaking—relies on indirect methods like immonium/oxonium ion screening. The authors propose integrating multiplexed tagging (e.g., tandem mass tags) with nDIA to further scale throughput. Emerging technologies, such as single-molecule protein sequencing, could complement MS-based SCP for even deeper insights.
The Chip-Tip workflow establishes a new paradigm for SCP, offering unparalleled sensitivity and scalability. By enabling precise PTM analysis and robust quantification of low-abundance proteins, it opens avenues for studying cellular dynamics in development, disease, and therapeutic responses. As multi-omics integration becomes mainstream, this technology will be pivotal in unraveling the molecular complexity of individual cells, ultimately advancing personalized medicine and systems biology.
Reference
- Ye, Z., Sabatier, P., van der Hoeven, L. et al. Enhanced sensitivity and scalability with a Chip-Tip workflow enables deep single-cell proteomics. Nat Methods 22, 499–509 (2025). https://doi.org/10.1038/s41592-024-02558-2
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

