CombPlex-a combinatorial staining platform coupled with an algorithmic framework
Current methods for imaging proteins in tissues, such as immunohistochemistry and immunofluorescence, are constrained by their reliance on dedicated measurement channels for each protein. This linear scalability inherently limits throughput, making proteome-scale imaging impractical. The study introduces CombPlex (Combinatorial Multiplexing), a novel framework that exponentially increases imaging capacity by combining combinatorial staining with deep learning-based decompression.
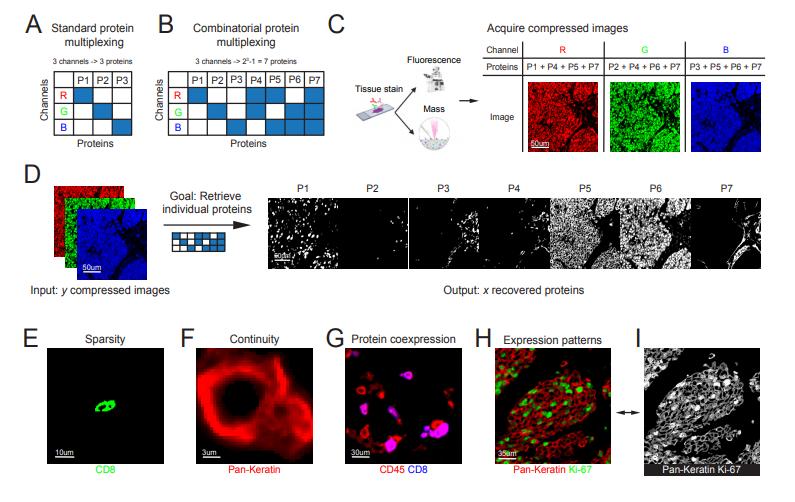
CombPlex employs a binary compression matrix 𝐴A, where each protein is assigned a unique combination of channels. For example, three channels can encode seven proteins. The compressed images, which aggregate signals from multiple proteins, are then decompressed using a two-step algorithm:
- Deep Learning Masking: A convolutional neural network (CNN) predicts binary masks identifying regions where each protein is expressed.
- Optimization: A least-squares optimization reconstructs pixel intensities using the masks as constraints.
This hybrid approach leverages spatial continuity, protein co-expression patterns, and subcellular localization-features learned implicitly by the CNN-to resolve underdetermined linear systems.
Key Results
- Simulations: Compressing 22 proteins into five channels yielded near-perfect reconstruction (F1 = 0.98, Pearson 𝑅=0.99R=0.99).
- Fluorescence Imaging (CODEX): Seven proteins compressed into three channels were reconstructed with high accuracy (F1 = 0.97, 𝑅=0.93R=0.93). Training on simulated data matched experimental performance, enabling flexibility.
- Mass-Based Imaging (MIBI): Six proteins compressed into three channels achieved robust results (F1 = 0.92, R=0.91R=0.91), even when trained on heterogeneous pre-existing datasets.
- Robustness: CombPlex tolerated missing proteins (up to 30% dropout) and variations in compression matrices. Errors primarily occurred at signal edges but did not compromise cell classification (balanced accuracy = 0.97–0.98).
Advantages and Implications
- Scalability: A 40-channel instrument could theoretically image 10121012 proteins, far exceeding current needs.
- Accessibility: Compatibility with standard fluorescence microscopes and mass spectrometers avoids costly hardware upgrades.
- Generalizability: Training on simulated or diverse datasets reduces reliance on paired experimental data, facilitating adoption.
CombPlex represents a paradigm shift in multiplexed imaging, decoupling throughput from channel limitations. By integrating combinatorial staining with deep learning, it enables proteome-scale spatial profiling without specialized instrumentation. Future advancements could extend its application to mRNA imaging or standardized clinical panels, revolutionizing pathology and systems biology. This work underscores the transformative potential of merging experimental design with machine learning to solve intractable biological challenges.
Reference
- Ben-Uri R, Ben Shabat L, Shainshein D, Bar-Tal O, Bussi Y, Maimon N, Keidar Haran T, Milo I, Goliand I, Addadi Y, Salame TM, Rochwarger A, Schürch CM, Bagon S, Elhanani O, Keren L. High-dimensional imaging using combinatorial channel multiplexing and deep learning. Nat Biotechnol. 2025 Mar 25. doi: 10.1038/s41587-025-02585-0. Epub ahead of print. PMID: 40133518.
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

