EVOLVEpro-a powerful tool for protein engineering
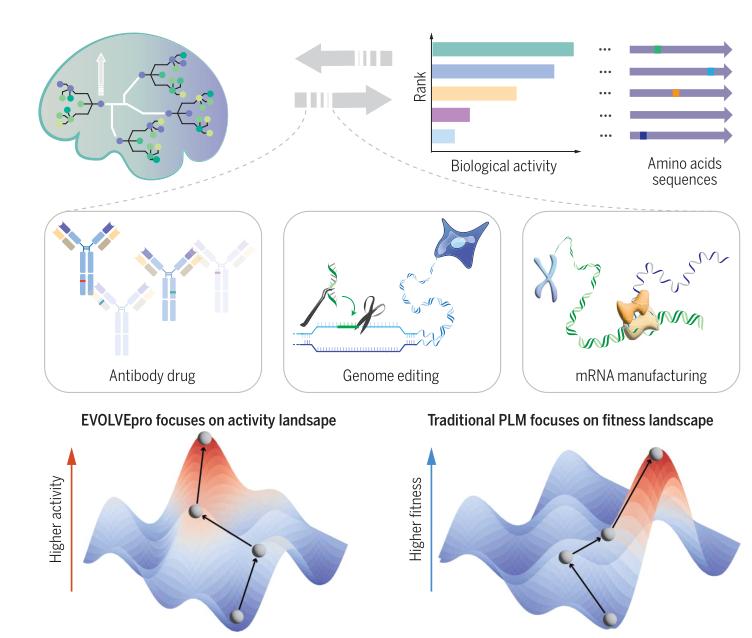
EVOLVEpro, a novel protein engineering framework that synergizes protein language models (PLMs) with few-shot active learning to optimize protein functions efficiently. Traditional directed evolution methods are labor-intensive and prone to local optima, while existing computational approaches, including zero-shot PLMs, struggle to generalize across diverse protein families or improve complex functionalities. EVOLVEpro addresses these limitations by integrating a regression model atop a PLM (ESM-2) and employing iterative experimental feedback to refine predictions, enabling rapid exploration of protein activity landscapes with minimal data.
Key innovations and results are:
- Architecture: EVOLVEpro combines ESM-2 embeddings (averaged across residues) with a lightweight random forest regressor. This setup learns protein activity landscapes through iterative rounds of experimental validation (as few as 10 data points per round), prioritizing high-activity mutants.
- Performance: Benchmarked across 12 deep mutational scanning datasets, EVOLVEpro outperformed zero-shot methods, achieving up to 40-fold improvements in antibody binding, 5-fold enhancements in CRISPR nuclease activity, and 100-fold increases in RNA polymerase fidelity.
- Multi-Objective Optimization: The framework successfully engineered proteins for conflicting objectives, such as balancing antibody binding affinity with expression yield or optimizing mRNA production for both high yield and low immunogenicity.
- Structural Insights: Mutations identified by EVOLVEpro often localized to functionally critical regions (e.g., DNA-binding domains in integrases or RNase H domains in reverse transcriptases), validated by AlphaFold-predicted structures. These mutations were frequently rare in natural sequences, underscoring the model's ability to explore novel mutational space.
- Divergence from PLM Fitness: EVOLVEpro's predictions showed little correlation with ESM-2's native fitness scores, emphasizing that evolutionary fitness (learned by PLMs) does not directly map to engineered activity. This highlights the necessity of experimental data to interpret PLMs effectively.
EVOLVEpro was applied to diverse proteins:
- Antibodies: Improved binding affinity for SARS-CoV-2 and transferrin receptor-targeting antibodies.
- Genome Editing Tools: Engineered compact Cas12f nucleases with enhanced activity for in vivo applications and optimized prime editors for large DNA insertions.
- RNA Production: Evolved T7 RNA polymerase variants producing mRNA with reduced immunogenic byproducts, enabling efficient circular RNA synthesis and improved in vivo gene expression.
The study underscores the limitations of relying solely on evolutionary fitness metrics from PLMs and advocates for hybrid approaches that integrate experimental feedback. EVOLVEpro's modular design allows compatibility with future advancements in generative PLMs or biophysical models, promising end-to-end protein design and optimization pipelines. By democratizing access to high-efficiency protein engineering with minimal experimental overhead, EVOLVEpro could accelerate therapeutic development, synthetic biology, and industrial enzyme design, bridging the gap between computational prediction and real-world functionality.
Reference
- Jiang K, Yan Z, Di Bernardo M, Sgrizzi SR, Villiger L, Kayabolen A, Kim BJ, Carscadden JK, Hiraizumi M, Nishimasu H, Gootenberg JS, Abudayyeh OO. Rapid in silico directed evolution by a protein language model with EVOLVEpro. Science. 2025 Jan 24;387(6732):eadr6006. doi: 10.1126/science.adr6006. Epub 2025 Jan 24. PMID: 39571002.
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

