Excitography, a novel protein purification method leveraging light-responsive molecular interactions
The technique utilizes a genetically encoded Azo-tag, a short peptide incorporating the non-canonical amino acid p-(phenylazo)-L-phenylalanine (Pap). Pap undergoes reversible trans-to-cis isomerization under UV light (355 nm), enabling precise control over protein binding and elution during affinity chromatography.
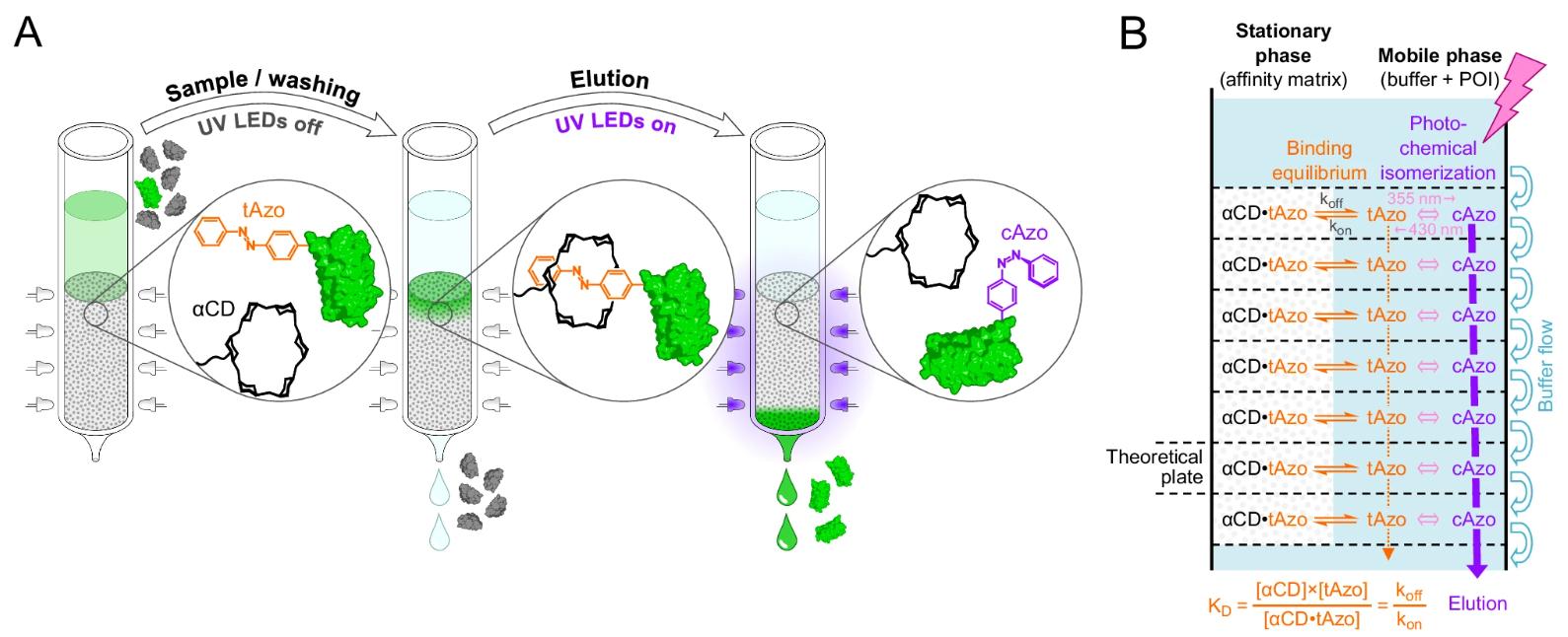
- Azo-tag Mechanism:
- In its trans state, Pap binds strongly to α-cyclodextrin (α-CD) immobilized on chromatography matrices (KD ≈ 91 μM).
- Upon UV irradiation (355 nm), Pap switches to the cis configuration, losing affinity for α-CD and enabling rapid, buffer-free elution.
- This eliminates the need for harsh elution agents (e.g., extreme pH, detergents) common in traditional affinity chromatography.
- Genetic and Strain Engineering:
- The authors engineered E. coli to site-specifically incorporate Pap into proteins using an evolved pyrrolysyl-tRNA synthetase (PyIRS) and a modified E. coli strain (NEBExpress(lowRF1)) with reduced release factor 1 (RF-1) activity to enhance amber codon suppression.
- Optimization of the Azo-tag's placement (N-terminal vs. C-terminal) and flanking residues (e.g., glycine spacers) improved binding efficiency and minimized truncation artifacts from incomplete amber suppression.
- Broad Applicability:
- Successful purification of diverse proteins, including enzymes (β-lactamase, glutathione-S-transferase), fluorescent proteins (mScarlet, sfGFP), and antibody fragments (scFv, nanobodies), validated the method's versatility.
- High-throughput compatibility was demonstrated using 96-well microtiter plates, enabling automated, parallel processing.
- Overcoming Challenges:
- Contaminants like E. coli maltose-binding protein (MBP) were mitigated by adding maltose or using an MBP-deficient strain.
- The Azo-tag's stability under reducing conditions and minimal impact on protein function were confirmed via enzymatic assays and structural analyses (CD spectroscopy).
- Advantages Over Existing Methods:
- Excitography avoids chemical elutants, preserving protein functionality and simplifying downstream applications.
- It outperforms other light-controlled systems (e.g., LOV2, phytochrome-based) by relying on a small, genetically encoded tag and robust α-CD interactions.
This work establishes Excitography as a scalable, gentle, and versatile purification platform. Its integration into industrial bioprocessing could streamline protein production for therapeutics and research. Future efforts may explore adapting the Azo-tag for in vivo applications or coupling it with optogenetic systems for dynamic protein regulation. The method's reliance on inexpensive LEDs and reusable α-CD matrices further enhances its practicality, promising broad adoption in biotechnology.
Reference
- Mayrhofer, P., Anneser, M.R., Schira, K. et al. Protein purification with light via a genetically encoded azobenzene side chain. Nat Commun 15, 10693 (2024). https://doi.org/10.1038/s41467-024-55212-y
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

