New technology for measuring the interaction between DNA and proteins
ORBIT (Origami-Rotor-Based Imaging and Tracking), a novel single-molecule method to track DNA rotation with millisecond resolution. Using a DNA origami rotor-a nanostructure with fluorescently labeled blades-ORBIT amplifies and visualizes rotational movements generated by DNA-processing enzymes. The rotor's design minimizes Brownian noise, enabling precise tracking of enzymatic activity. Applied to RecBCD helicase (involved in DNA repair), ORBIT revealed ATP-independent initiation phases, processive unwinding (~300 bp/s), backtracking, and substrate-dependent mechanisms. For RNA polymerase (RNAP), ORBIT captured single-base-pair rotational steps (~35°) during transcription, resolving dynamics previously undetectable without applied force. The method's high throughput and compatibility with standard microscopes position it as a transformative tool for studying enzyme mechanics and DNA-protein interactions.
Understanding how enzymes manipulate DNA-such as unwinding, transcription, or repair-requires observing rotational dynamics at unprecedented resolution. Traditional methods like optical tweezers or magnetic torque spectroscopy face limitations in throughput, temporal resolution, or reliance on applied forces. The ORBIT method, detailed in this study, overcomes these barriers by leveraging DNA nanotechnology to illuminate enzyme mechanics with unparalleled clarity.
ORBIT's core innovation lies in its DNA origami rotor, a precisely engineered nanostructure that amplifies rotational motion. The rotor's blades, extending 80 nm from a central axis, magnify DNA twists while minimizing hydrodynamic drag. Fluorescent tags on the blades enable real-time tracking via total internal reflection fluorescence (TIRF) microscopy, achieving ~10 nm spatial and 1 ms temporal resolution. By anchoring enzymes (e.g., RecBCD or RNAP) to a surface and attaching DNA substrates to the rotor, ORBIT converts rotational forces into measurable fluorescent trajectories. This design bypasses the need for external forces, offering a "torque-free" platform that preserves physiological conditions.
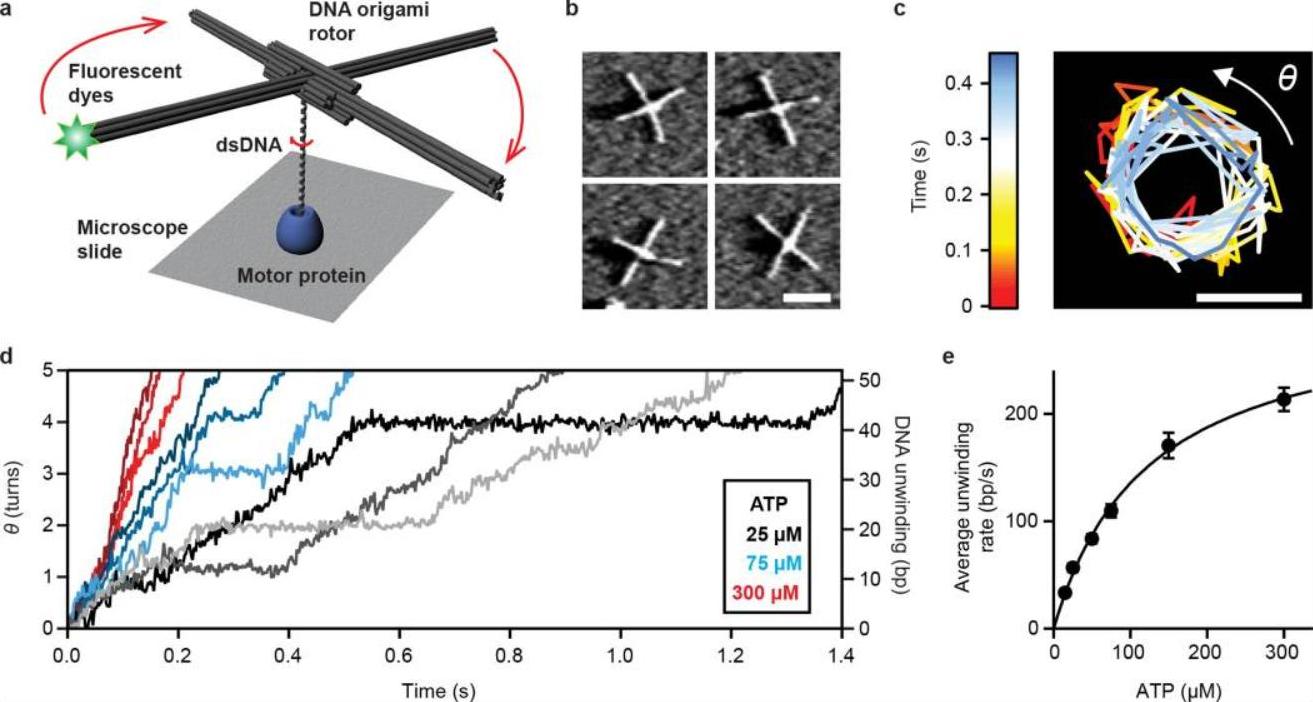
RecBCD, critical for repairing double-strand breaks, unwinds DNA via coordinated motor domains. ORBIT revealed previously hidden initiation mechanisms:
- ATP-Independent Unwinding: RecBCD transiently unwinds ~5 bp without ATP, creating a "gateway" to engage its RecB motor. This step is absent in substrates with pre-existing 3′ overhangs, highlighting substrate-specific initiation strategies.
- Pausing and Backtracking: Even without opposing force, RecBCD exhibits pauses and occasional backtracking (~6 bp), with recovery times dependent on ATP concentration. These dynamics suggest a kinetic "tug-of-war" between motor subunits.
- Substrate Geometry Matters: DNA ends with 5′ overhangs prolong initiation, whereas weakening base-pairing (e.g., replacing G-C with A-T) accelerates it, underscoring sequence-dependent regulation.
For RNAP, ORBIT resolved rotational steps corresponding to single-base-pair translocation (~35°), aligning with B-DNA's helical twist. These steps, observed at low NTP concentrations, demonstrate tight coupling between RNAP's movement and DNA's helical geometry. The absence of applied force in these measurements challenges prior assumptions that force is necessary to detect such fine-scale dynamics.
ORBIT's modularity—adjustable rotor size, compatibility with diverse enzymes, and minimal instrumentation requirements—positions it as a versatile tool. Potential applications include:
- High-Throughput Drug Screening: Testing inhibitors targeting helicases or polymerases.
- Nanoscale Actuators: Integrating rotors with synthetic molecular machines.
- Multi-Enzyme Studies: Observing coordination between enzymes during replication or repair.
By bridging the gap between structural biology and real-time enzymology, ORBIT opens new frontiers in understanding life's molecular machinery. Its success underscores the power of DNA nanotechnology to transform biophysical inquiry, offering a lens through which the invisible dynamics of life's processes become vividly clear.
Reference
- Kosuri P, Altheimer BD, Dai M, Yin P, Zhuang X. Rotation tracking of genome-processing enzymes using DNA origami rotors. Nature. 2019 Aug;572(7767):136-140. doi: 10.1038/s41586-019-1397-7. Epub 2019 Jul 17. PMID: 31316204; PMCID: PMC7036295.
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

