paDSF-Protein-Adaptive Differential Scanning Fluorimetry and the Aurora Dye Library
Differential Scanning Fluorimetry (DSF) has long been a cornerstone technique for studying protein stability, ligand binding, and structural dynamics by monitoring thermal unfolding through fluorescent dyes. However, its utility has been constrained by the limited compatibility of conventional dyes like SYPRO Orange with diverse proteins, failing in over 70% of cases. A groundbreaking study introduces protein-adaptive DSF (paDSF)-a transformative approach that overcomes these limitations through a curated library of 312 chemically diverse dyes, termed the Aurora library. This innovation not only triples DSF's success rate but also expands its applications to complex biochemical phenomena.
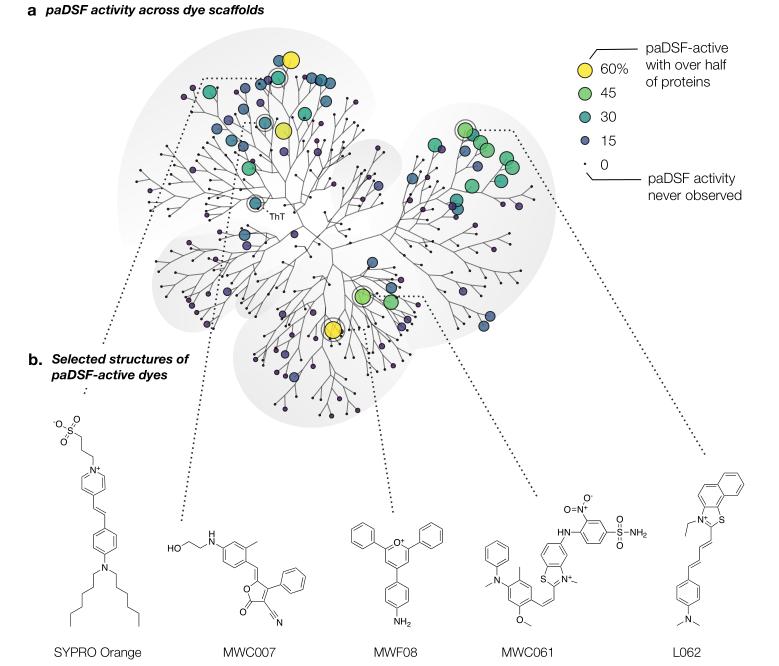
The Aurora library reimagines DSF by sourcing dyes from unconventional domains, including textile, histology, and laser industries. These dyes were rigorously screened against 60 biochemically diverse proteins, ranging from small disordered peptides to large multidomain enzymes. By employing counter-screens to eliminate dyes with nonspecific fluorescence or buffer incompatibilities, researchers identified 107 "paDSF-active" dyes, with 48 high-performing candidates forming the streamlined Aurora-concise subset. Remarkably, 94% of tested proteins (66/70)—including 10 SARS-CoV-2 viral proteins—were successfully analyzed, a stark contrast to the 29% success rate of SYPRO Orange alone. Each protein paired with an average of 13 dyes, highlighting the library's versatility.
Practical Applications: From Viral Targets to Allostery
- SARS-CoV-2 Protein Studies:
paDSF enabled rapid assay development for critical SARS-CoV-2 proteins involved in viral replication, such as PLPro and the nsp3 macrodomain. By replacing SYPRO Orange with dye T004, researchers reduced compound interference in small-molecule screens from 8% to 2%, facilitating the discovery of high-affinity inhibitors. The method's spectral diversity (detection across blue to near-infrared wavelengths) further minimizes experimental artifacts. - Unraveling Interdomain Allostery:
The multidomain enzyme O-GlcNAc Transferase (OGT) showcased paDSF's ability to monitor complex conformational changes. Dye TW408 revealed two distinct thermal transitions corresponding to OGT's catalytic and client-binding domains. Titration of the allosteric ligand L4 induced opposing shifts in these transitions, illustrating domain coupling—a phenomenon undetectable with traditional DSF. This breakthrough underscores paDSF's capacity to resolve intricate biochemical interactions.
Chemoinformatic analyses revealed that paDSF dyes operate through mechanisms distinct from simple hydrophobic interactions. Hydrophilic dyes like MWC007 performed exceptionally, suggesting fluorescence responsiveness relies on structural motifs or specific protein-dye interactions. Each protein exhibited a unique "dye fingerprint," with selectivity ranging from single-protein specificity to broad compatibility. This diversity implies that paDSF dyes detect varied features of unfolded states, such as exposed residues or conformational flexibility.
paDSF's workflow requires only 100 μL of low-concentration protein (5 μM) and completes screening within hours, making it accessible for high-throughput studies. Its resilience to common additives (detergents, reducing agents) and compatibility with disordered or phase-separated proteins further broadens its scope. Future efforts could explore dye design principles to target specific protein features or integrate paDSF with advanced denaturation protocols for deeper mechanistic insights.
By reframing the dye as a tunable variable, paDSF transcends the constraints of traditional DSF, emerging as a universal platform for studying protein dynamics. Its success in analyzing SARS-CoV-2 targets and capturing allostery in OGT exemplifies its transformative potential in drug discovery and structural biology. As the Aurora library opens new avenues for probing protein behavior, paDSF stands poised to become an indispensable tool in the biochemist's arsenal, bridging the gap between simplicity and versatility in experimental design.
Reference
- Wu T, Yu JC, Suresh A, Gale-Day ZJ, Alteen MG, Woo AS, Millbern Z, Johnson OT, Carroll EC, Partch CL, Fourches D, Vinueza NR, Vocadlo DJ, Gestwicki JE. Conformationally responsive dyes enable protein-adaptive differential scanning fluorimetry. bioRxiv [Preprint]. 2023 Jan 28:2023.01.23.525251. doi: 10.1101/2023.01.23.525251. Update in: Nat Biotechnol. 2025 Jan;43(1):106-113. doi: 10.1038/s41587-024-02158-7. PMID: 36747624; PMCID: PMC9900766.
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

