SYMPLEX-designed to mine biomedical literature and databases for synthetic biology parts
The rapid development of mRNA vaccines during the COVID-19 pandemic highlighted the importance of efficient mRNA capping, a process critical for mRNA stability and translational efficacy. However, reliance on a single commercial enzyme-the vaccinia virus-derived capping enzyme-has posed limitations in scalability and cost. This study addresses this challenge by leveraging AI to mine nature's vast genetic diversity for superior alternatives.
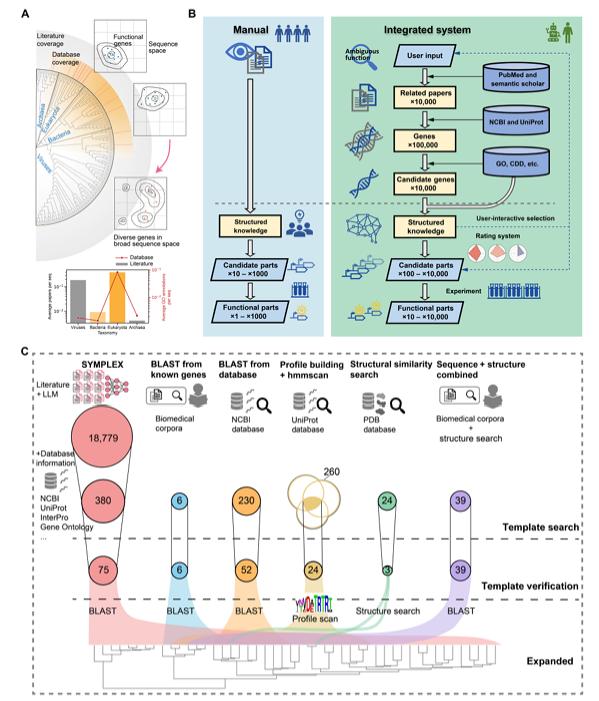
SYMPLEX operates through three stages:
- Literature Retrieval: Using LLMs, the platform identifies relevant papers and extracts gene-function-species relationships from unstructured text.
- Knowledge Integration: Extracted data are mapped to standardized biological ontologies (e.g., Gene Ontology terms) and cross-referenced with databases like UniProt and NCBI.
- Candidate Prioritization: Genes are scored based on textual evidence, domain annotations, and evolutionary diversity, enabling the selection of high-potential candidates.
This approach overcomes the "homology trap" of traditional bioinformatics by uncovering distantly related or entirely novel enzymes missed by sequence-based searches.
- Diverse CEs with Biotechnological Promise: SYMPLEX identified CEs from viruses, eukaryotes, and mobile genetic elements. For example, Marseillevirus-derived enzymes (MRV_5/6) exhibited superior catalytic efficiency, while compact viral CEs demonstrated modular domain arrangements suitable for engineering.
- Structural Insights: AlphaFold2-predicted structures revealed conserved catalytic cores and flexible peripheral domains, providing a blueprint for rational enzyme design.
- Validation Across Systems: In vivo tests in yeast and mammalian cells confirmed cross-species functionality, a critical feature for industrial applications.
SYMPLEX represents a leap forward in AI-driven bioengineering. By automating literature mining and integrating multimodal data, the platform accelerates the discovery of functional genetic parts. This approach is not limited to CEs; the authors demonstrate its applicability to other enzymes, such as glutathione S-transferases, suggesting broad utility in metabolic engineering and drug development.
Broader Impact
- mRNA Technology: The discovery of efficient, compact CEs could reduce production costs for mRNA vaccines and therapies.
- Green Chemistry: Enzymes like MRV_5/6 may enable sustainable synthesis of biomaterials, such as biodegradable plastics.
- AI in Life Sciences: SYMPLEX exemplifies how LLMs can transform biological research, turning fragmented literature into actionable knowledge.
While SYMPLEX significantly reduces noise, challenges remain, including gaps in database annotations and the need for full-text access to older publications. Future iterations could incorporate generative AI for de novo enzyme design or predict functional synergy between mined parts.
The integration of AI with synthetic biology heralds a new era of rapid, data-driven innovation. SYMPLEX's success in discovering high-performance CEs demonstrates the untapped potential of biological diversity and the power of machine learning to unlock it. As AI models evolve, platforms like SYMPLEX will become indispensable tools for addressing global health and environmental challenges, paving the way for a future where biotechnology is limited only by imagination.
Reference
- Wang T, Qin BR, Li S, Wang Z, Li X, Jiang Y, Qin C, Ouyang Q, Lou C, Qian L. Discovery of diverse and high-quality mRNA capping enzymes through a language model-enabled platform. Sci Adv. 2025 Apr 11;11(15):eadt0402. doi: 10.1126/sciadv.adt0402. Epub 2025 Apr 9. PMID: 40203090; PMCID: PMC11980835.
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

