DenseMAP-Space time specific photocrosslinking technology for efficient and universal protein interactions
In recent years, the liquid-liquid phase separation (LLPS) phenomenon of biomolecules and the resulting membraneless subcellular structure - biomolecular condensates - have become a hot topic in the field of scientific research. These aggregates can spatially separate biomolecules without relying on membrane structure, thereby achieving precise spatiotemporal regulation of cellular physiological processes. Among them, protein-protein interaction (PPI) is an important basis for this regulatory mechanism. Therefore, in-depth analysis of protein interactions in specific biomolecule aggregates is of great significance for revealing their formation mechanisms and functions.
However, due to the dynamic formation process of biomolecule aggregates and the lack of clear membrane structure boundaries, traditional methods such as physical separation and immunoprecipitation are difficult to effectively preserve their interaction information. Meanwhile, the complexity and crowding of its internal structure make the proximity labeling method based on diffusible probes prone to producing many false positive results. Unnatural amino acids with photo crosslinking groups can be integrated into proteins of interest (POI) during protein translation and generate active intermediates fixed on the protein upon photoexcitation, thereby achieving highly spatiotemporal specific crosslinking and widely used for capturing direct and transient PPIs in living cells. Among them, metabolically insertable photo crosslinked amino acids, as analogues of natural amino acids, can globally introduce corresponding amino acid sites into the entire proteome through cellular metabolic pathways, achieving multi-point insertions and having advantages in capturing multivalent interactions in LLPS systems.
DenseMAP (Condensation-enhanced Spatial-directed Metabolic Incorporation-Assisted Photo-crosslinking) efficiently captures and identifies protein interactions of specific biomolecule aggregates in living cells by combining spatially specific protein labeling with enhanced photocrosslinking.
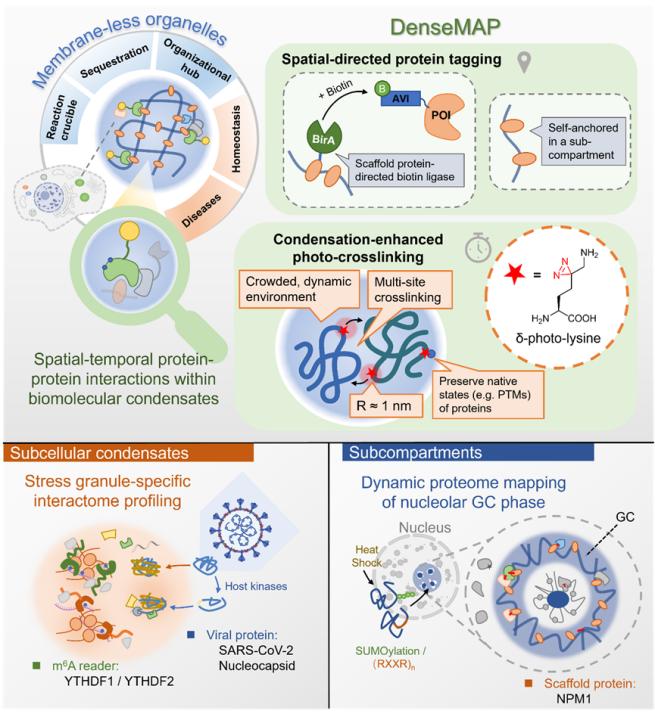
Firstly, the research team used bioinformatics methods to analyze the distribution of 20 amino acids in the protein assembly of biomolecules. They found that lysine had a high abundance and was more distributed at the protein interaction interface due to the positive residue band. Based on this, they developed a new type of photo crosslinked lysine (δ - photo lysine, δ K *) with a photo crosslinking group called diazirine at the δ - position. Compared with the previously reported photo crosslinked lysine with a photo crosslinking group at the gamma position, the new probe is easier to synthesize and has higher photo crosslinking efficiency. They also verified that the insertion of δ K * had little effect on the normal physiological state and protein function of cells. Later, they demonstrated through cross-linking experiments with 1,6-hexanediol treatment and phase separation inactivated mutants that based on the nanoscale cross-linking radius and multi-point insertion characteristics, δ K * has higher cross-linking efficiency in crowded phase separation systems, achieving a "coagulation enhancement" effect.
Due to the complex organizational structure of biomolecule aggregates, proteins can be roughly divided into two categories: "client" proteins and "scaffold" proteins. The former is generally distributed in multiple subcellular regions and only enters dynamically formed biomolecule aggregates under specific conditions; The latter mediates the formation of specific biomolecule aggregates, often with a single spatial localization, and their multivalent interactions generally cover the "client" proteins within them. Therefore, for different types of proteins, the research team designed two application routes for DenseMAP.
For "client" proteins with multiple locations simultaneously, the research team labeled them with the "skeleton" proteins of specific biomolecule aggregates coupled with biotin ligase (BirA), which cooperated with DenseMAP's coagulation enhancement cross-linking to specifically capture the interacting proteome of POIs located within the interested biomolecule aggregates. Based on this approach, the interaction proteome of SARS-CoV-2 nucleocapsid (N) protein in stress granules (SG) was captured and identified using G3BP1 BirA binding to δ K *. The promotion effect of host kinase mediated phosphorylation on the interaction of N protein with many functional RNA binding proteins in SG was analyzed, further elucidating the mechanism of virus host antagonism. They also identified and compared the interaction proteome of m6A modified "readers" YTHDF1 and YTHDF2 in SG, verifying previous conclusions that the two have different properties during SG formation.
On the other hand, due to the complex "subcompartment" structure exhibited by many biomolecule aggregates, each subcompartment often maintains its structure and recruits "customer" proteins through specific "skeleton" proteins, thereby achieving dynamic exchange of substances. However, due to their tightly interwoven structure, the proteome of these subregions is difficult to accurately distinguish and identify through traditional physical separation or proximity labeling methods. It is worth noting that for single localized "skeleton" proteins, direct interacting proteins in specific subregions can be obtained without the need for additional labeling methods. Due to their extensive direct interactions with "client" proteins in the region, the interaction group of these proteins can actually be seen as a microcosm of the protein group in that subregion. Based on this, the use of DenseMAP strategy to obtain the interaction group of "skeleton" proteins provides a new perspective for in-depth study of the properties of specific subregions in biological molecular aggregates.
Previous studies have shown that the outermost granular component (GC) layer of the nucleolus can store misfolded proteins in the nucleus under pressure conditions, thereby protecting cells from maintaining normal function and playing a key role in protein quality control (PQC). In order to further explore this mechanism, the research team used the DenseMAP strategy to capture and identify the interaction group of the "skeleton" protein NPM1 in the GC layer under normal physiological conditions and thermal stimulation conditions. It was found that after thermal stimulation, the content of SUMO modified proteins in the GC sublayer proteome significantly increased. After a series of biological verifications, the research team revealed that SUMOylation modifications, especially SUMO2/3, can bind to NPM1 through its R-motif on the sequence, and recruit misfolded proteins that do not contain R-motif themselves into the nucleolus for temporary storage. This discovery not only provides important supplements to the mechanism of nucleoli in PQC, but also offers new clues and evidence for understanding the pathogenesis of neurodegenerative diseases such as ALS and exploring potential therapeutic strategies.
In summary, this study cleverly integrated spatially specific protein labeling technology with condensed enhanced metabolic photocrosslinking probes, and developed an efficient and universal spatiotemporal specific photocrosslinking technology - DenseMAP. This strategy can accurately capture and identify the protein-protein interactions within specific biomolecule aggregates in living cells, providing a powerful tool for further elucidating the functional mechanisms, dynamic changes, and disease-related mechanisms of membraneless organelles.
Reference
- Li, K., Xie, X., Gao, R. et al. Spatiotemporal protein interactome profiling through condensation-enhanced photocrosslinking. Nat. Chem. 17, 111–123 (2025). https://doi.org/10.1038/s41557-024-01663-1
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

