Frizzled Receptors
Related Symbol Search List
Immunology Background
About Frizzled Receptors
Frizzled (FZD) family proteins belong to the G-protein coupled receptors (GPCRs), which are the largest group of cell surface receptors found in humans. There are six classes of GPCRs: Rhodopsin family (Class A), secretin receptor family (Class B), metabotropic glutamate/pheromone family (Class C), fungal mating pheromone receptor family (Class D), cyclic AMP receptor family (Class E), and FZD/Smoothened (SMO) family (Class F). Compared to other GPCRs, FZD family receptors are thought to be atypical as the activity level of G protein signaling mediated by FZD is much lower than that of a typical GPCR. Spontaneous and targeted mutations in mammalian Frizzled genes have revealed their essential roles in various developmental and homeostatic processes, including palate and heart morphogenesis, tissue polarity, vascular formation, and maintenance, and the development of the central nervous system, kidney, and bone. Increasing evidence also suggests that FZDs play an important role in cancer development and progression.
There are ten members in the Frizzled family, FZD1-10 in humans and Fzd1-10 in mice. They are between 500~700 amino acids in length and can be divided into four sub-families based on the amino acid sequence homology. Common structures of FZDs include an extracellular N-terminus, seven hydrophobic transmembrane domains, and an intracellular C-terminus. The N-terminus contains a conserved 120 amino acids cysteine-rich domain (CRD) connected to the first transmembrane helix by a hydrophilic linker of 70-120 amino acids. FZD utilizes CRD as a necessary and sufficient binding site for various ligands, including the WNT proteins, R-spondin, and Frizzled-related proteins.
Mechanism of Action of Frizzled Receptors
Frizzled receptors can activate three signaling pathways: canonical WNT/β-catenin pathway, non-canonical planar cell polarity (PCP) pathway, and WNT/calcium signaling pathway. The canonical WNT/β-catenin signaling pathway is characterized by the stabilization of β-catenin upon ligand binding. The PCP pathway controls the cell/tissue polarity along the body axis. When mutated, results in various developmental defects in the body. The WNT/calcium signaling pathway is defined by the regulation of intracellular Ca2+ levels and the activation of many calcium-sensitive enzymes. Although the activation of these three signaling pathways seems distinct and independent, convergent models of signaling networks where several pathways act in a coordinated and interdependent manner have been proposed.
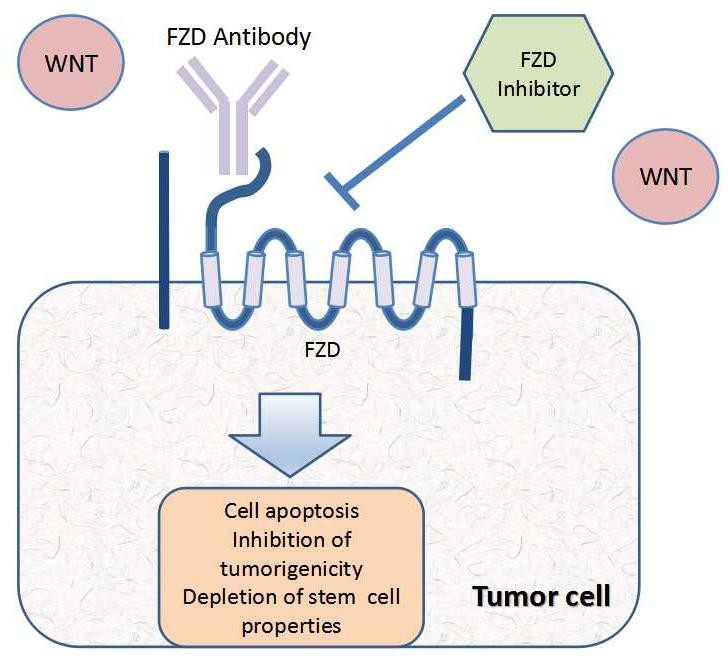 Fig.1 Wnt signalling pathways. A) FZD/WNT/β-catenin signalling. B) FZD/WNT/PCP signalling. C) FZD/WNT/Ca2+ signalling.
Fig.1 Wnt signalling pathways. A) FZD/WNT/β-catenin signalling. B) FZD/WNT/PCP signalling. C) FZD/WNT/Ca2+ signalling.
The mechanism of action of Frizzled receptors involves the following key steps:
- Wnt binding and Activation: Frizzled receptors are key components of the Wnt signaling pathway and initiate signaling by binding to Wnt ligands, a class of secreted glycoproteins that trigger a series of intracellular responses upon binding to frizzled receptors.
- Co-Receptor Involvement: Frizzled receptors usually require interaction with co-receptors (e.g. LRP5/6) to initiate signaling. binding of Wnt ligands to frizzled receptors and co-receptors triggers a conformational change in the frizzled receptor, which activates intracellular signaling pathways.
- Downstream Signaling: Activation of frizzled receptors triggers a series of downstream signaling events. In the classical Wnt/β-catenin pathway, activated frizzled receptors lead to stabilization and nuclear translocation of β-catenin, which in turn regulates gene expression. In the non-classical pathway, frizzled receptors regulate processes such as cytoskeleton construction, cell migration, and tissue polarity by activating small GTPases and other downstream effector molecules.
Functions of Frizzled Receptors
Frizzled receptors play important functions in several biological processes, including:
- Embryonic Development
Frizzled receptors play key roles in embryonic development, including axial formation, cell fate determination, and tissue pattern formation. They are involved in embryonic orientation and organ formation and are essential for normal embryonic development.
- Tissue Homeostasis and Regeneration
Frizzled receptors play a key role in tissue homeostasis and regeneration in adult organisms. They regulate cell proliferation, differentiation, and stem cell maintenance, and are important for maintaining normal tissue function and repairing damaged tissues.
- Neurodevelopment and Neurological Disorders
Frizzled receptors are involved in neurodevelopmental processes such as neuronal migration, axon guidance, synapse formation, etc. Abnormal regulation of the Frizzled signaling pathway has been linked to the onset and progression of neurological disorders such as Alzheimer's disease and autism spectrum disorders.
- Cancer and Metastasis
The aberrant Frizzled receptor signaling pathway is associated with cancer progression and metastasis. Frizzled receptors promote tumor growth, invasion, and angiogenesis, and play a key role in tumor formation and metastasis by regulating processes such as cell proliferation, migration, and epithelial-mesenchymal transition.
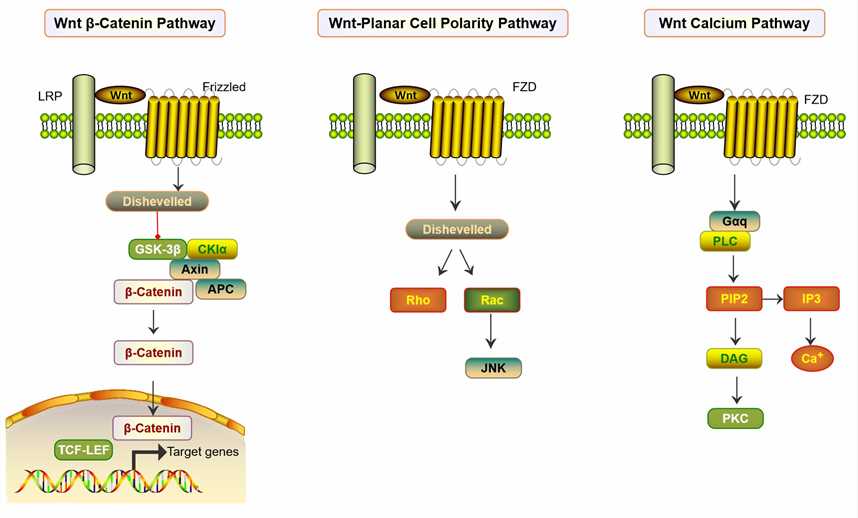 Fig.2 Frizzled Receptors as Potential Therapeutic Targets in Human Cancers. (Zeng CM, et al., 2018)
Fig.2 Frizzled Receptors as Potential Therapeutic Targets in Human Cancers. (Zeng CM, et al., 2018)
Available Resources for Frizzled Receptors
The in-depth study of the mechanism of action and function of Frizzled receptors is important for understanding the Wnt signaling pathway, developmental biology, and the mechanisms of related diseases. Creative BioMart offers products and services related to Frizzled receptors, providing researchers with valuable tools and resources to further explore the mechanism of action and function of these receptors. The following frizzled receptors are displayed, click to view all related molecules/targets and research reagents. Please feel free to contact us with any questions or requests.
References:
- Wang Y, Chang H, Rattner A, Nathans J. Frizzled Receptors in Development and Disease. Curr Top Dev Biol. 2016;117:113-139.
- Alrefaei A F. Frizzled receptors (FZD) play multiple cellular roles in development, in diseases, and as potential therapeutic targets - ScienceDirect[J].Journal of King Saud University - Science, 2021.
- Zeng CM, Chen Z, Fu L. Frizzled Receptors as Potential Therapeutic Targets in Human Cancers. Int J Mol Sci. 2018;19(5):1543.

