Myeloid Cell
Related Symbol Search List
Immunology Background
About Myeloid Cell
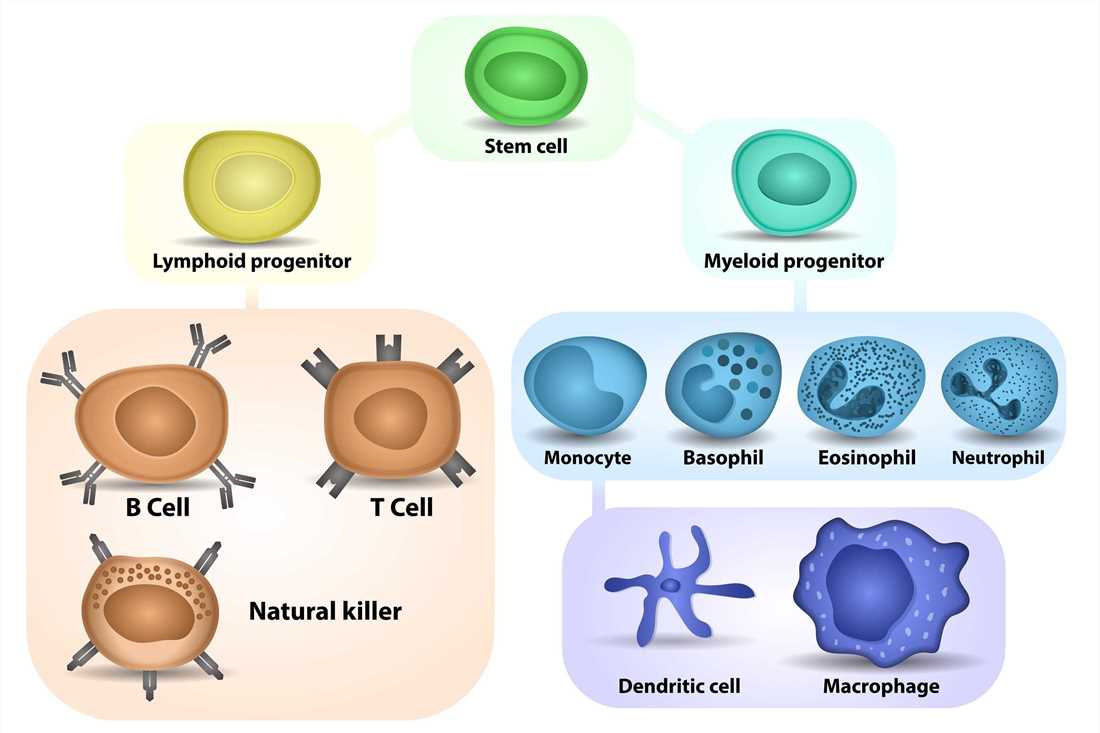
Granulocytes, monocytes, macrophages, and dendritic cells (DCs) represent a subgroup of leukocytes, collectively called myeloid cells. They circulate through the blood and lymphatic system and are rapidly recruited to sites of tissue damage and infection via various chemokine receptors. Within the tissues they are activated for phagocytosis as well as secretion of inflammatory cytokines, thereby playing major roles in protective immunity. Myeloid cells can also be found in tissues under steady-state conditions, where they control development, homeostasis, and tissue repair. Genetic alterations in myeloid cells may cause an abnormal increase in mature myeloid or blast cells resulting in chronic or acute myelogenous. Here is an introduction to some key myeloid cell types:
- Neutrophils: Neutrophils are the most abundant type of white blood cells and are often the first responders to infections. They are highly mobile and phagocytic, capable of engulfing and destroying bacteria, fungi, and other pathogens. Neutrophils are characterized by their multilobed nucleus and granules containing antimicrobial molecules.
- Monocytes/Macrophages: Monocytes are circulating immune cells that can differentiate into tissue-resident macrophages or dendritic cells upon migration into tissues. Macrophages are phagocytic cells involved in immune defense, tissue homeostasis, and wound healing. They engulf and eliminate pathogens, cellular debris, and dead cells, and also produce cytokines and chemokines that regulate immune responses.
- Dendritic Cells: Dendritic cells are specialized antigen-presenting cells that capture, process, and present antigens to T cells, initiating adaptive immune responses. They are highly efficient at antigen uptake and have unique cell surface receptors that enable them to recognize pathogens and activate T cells.
- Eosinophils: Eosinophils are involved in allergic reactions and defense against certain parasites. They release toxic substances to destroy parasites and modulate inflammation. Eosinophils also play a role in asthma, allergic rhinitis, and other eosinophilic disorders.
- Basophils: Basophils are involved in allergic reactions and immune responses against parasites. They release histamine and other mediators, contributing to inflammation and allergic symptoms. Basophils also play a role in chronic inflammatory conditions such as asthma and chronic urticaria.
- Mast Cells: Mast cells are tissue-resident cells found throughout the body, particularly in connective tissues and mucosal surfaces. They are involved in allergic reactions and immune responses against pathogens. Mast cells release histamine, cytokines, and other mediators, contributing to inflammation and allergic symptoms.
These are just a few examples of myeloid cells, and there are other subsets and specialized populations within the myeloid lineage. They interact with other immune cells, orchestrate immune responses, and contribute to the overall functioning of the immune system.
Myeloid Cell Related Diseases
There are several diseases and disorders associated with myeloid cell dysfunction. Here are some examples:
- Leukemia: Leukemia is a type of cancer that affects the bone marrow and results in the abnormal proliferation of myeloid cells. Myeloid leukemia can arise from different stages of myeloid cell development, such as acute myeloid leukemia (AML) or chronic myeloid leukemia (CML). These conditions lead to an overproduction of immature or abnormal myeloid cells, which can interfere with normal blood cell production and function.
- Myeloproliferative Neoplasms (MPNs): MPNs are a group of clonal disorders characterized by the overproduction of mature myeloid cells in the bone marrow. Examples of MPNs include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). These conditions can lead to an abnormal increase in red blood cells, platelets, or fibrous tissue in the bone marrow, causing various symptoms and complications.
- Myelodysplastic Syndromes (MDS): MDS is a group of disorders characterized by abnormal production and maturation of myeloid cells in the bone marrow. In MDS, the bone marrow produces immature or dysfunctional myeloid cells that do not fully mature into healthy blood cells. This can lead to low blood cell counts (cytopenias) and an increased risk of developing AML.
- Chronic Myelomonocytic Leukemia (CMML): CMML is a rare type of leukemia characterized by the presence of both increased monocytes and abnormal myeloid cells in the bone marrow and blood. It is considered a myelodysplastic/myeloproliferative neoplasm (MDS/MPN) overlap disorder, as it shares features of both MDS and MPNs.
- Hereditary Neutrophil Disorders: There are inherited disorders that affect neutrophils, such as chronic granulomatous disease (CGD) and leukocyte adhesion deficiency (LAD). CGD is a primary immunodeficiency characterized by defective neutrophil function, leading to recurrent bacterial and fungal infections. LAD is a rare genetic disorder that impairs the ability of neutrophils to migrate and adhere to the site of infection, resulting in recurrent infections and delayed wound healing.
These are just a few examples of diseases associated with myeloid cell dysfunction. Many other conditions, including autoimmune disorders, inflammatory diseases, and certain infections, can involve dysregulation or abnormal function of myeloid cells. Precise diagnosis, treatment, and management of these conditions often require a comprehensive understanding of myeloid cell biology and their role in immune responses.
Available Resources for Myeloid Cell
Creative BioMart offers a wide range of quality tools for myeloid cell research, including recombinant proteins and more. We provide personalized services according to the specific needs of our clients. In addition, we provide comprehensive resources covering all aspects of myeloid cells, including related pathways, protein functions, interacting proteins, related articles, and other relevant topics to help advance myeloid cell research.
Our Featured Products
- Active Recombinant Human ANPEP, His tagged
- Recombinant Mouse Anpep, His tagged
- Recombinant Human CD163, GST-tagged
- Active Recombinant Human CD33 Protein, Fc-tagged, Alexa Fluor 488 conjugated
- Active Recombinant Human CD33 Protein, Fc-tagged, FITC conjugated
- Recombinant Human CD33 Molecule, Fc Chimera
- Recombinant Human CD33 Protein, Fc-tagged, Alexa Fluor 555 conjugated
- Recombinant Mouse Cd33 Protein, His-tagged, Alexa Fluor 647 conjugated
- Recombinant Human CD68, Fc-His tagged
- Recombinant Mouse Cd68 protein, His-tagged
- Recombinant Human CD14 protein, hFc-tagged
- Recombinant Mouse Cd14, His tagged
- Recombinant Human BST1 Protein, His-tagged
- Recombinant Mouse Bst1, His tagged
- Active Recombinant Human CD34 protein, His-tagged
- Recombinant Mouse Cd34, His tagged
- Recombinant Human CEACAM8 protein, His-tagged
- Active Recombinant Human CEACAM8 protein, His-Avi-tagged, Biotinylated
- Recombinant Human SIGLEC5 Protein, hIgG-His-Tagged
If you have any questions, requirements, or cooperation intentions, please feel free to contact us. We very much look forward to working with you and helping you achieve research and commercial success.
References:
- Kawamoto H, Minato N. Myeloid cells. Int J Biochem Cell Biol. 2004;36(8):1374-1379. doi:10.1016/j.biocel.2004.01.020
- Weiskopf K, Schnorr PJ, Pang WW, et al. Myeloid Cell Origins, Differentiation, and Clinical Implications. Microbiol Spectr. 2016;4(5):10.1128/ microbiolspec. MCHD-0031-2016. doi:10.1128/microbiolspec.MCHD-0031-2016

