Cytotoxic T Cells (CTLs)
Related Symbol Search List
Immunology Background
Available Resources Related to Cytotoxic T Cells (CTLs)s
Creative BioMart offers a wide range of products, customized services, and resources related to cytotoxic T cells to meet all of your CTLs research needs, designed to accelerate your research and contribute to advances in the field of cytotoxic T cells.
We offer a variety of high-quality products designed to support CTL research. Our portfolio includes antibodies, recombinant proteins, protein-coupled magnetic beads, cell and tissue lysates, chromatography reagents, assay kits, and others. |
We understand the unique needs of CTLs-related researchers. Our team of experts is ready to provide tailored solutions through our customized services. |
We offer a wide range of resources related to CTLs, including protein functions, protein interactions, scientific literature, and other valuable information, providing researchers with strong support to help advance their research programs and experiments. |
About Cytotoxic T Cells (CTLs)
Cytotoxic T cells (CTLs), also known as CD8+ T cells, are a critical component of the adaptive immune system. They play a crucial role in eliminating infected or abnormal cells, including virus-infected cells, cancer cells, and cells presenting foreign antigens. CTLs are specifically designed to recognize and destroy target cells through a process called cell-mediated cytotoxicity. Here is an introduction to the key features and functions of CTLs:
- Development and Activation: CTLs are derived from precursor cells in the bone marrow. They undergo maturation and selection processes in the thymus, where they acquire their T-cell receptors (TCRs) and develop specificity for a particular antigen. Upon encountering their specific antigen, usually presented by major histocompatibility complex class I (MHC-I) molecules, CTLs become activated and undergo clonal expansion to generate a large population of effector CTLs.
- TCR-MHC Interaction: The TCR on CTLs recognizes antigenic peptides presented by MHC-I molecules on the surface of target cells. This interaction triggers the activation of CTLs and initiates the effector functions of CTLs against the target cells. The TCR-MHC interaction is highly specific, enabling CTLs to distinguish between healthy and infected/abnormal cells.
- Effector Functions: Once activated, CTLs employ various effector mechanisms to eliminate target cells. These mechanisms include: a. Perforin-Granzyme Pathway: CTLs release perforin and granzymes, which are cytotoxic molecules. Perforin forms pores in the target cell membrane, allowing granzymes to enter the target cell. Granzymes then induce apoptosis, leading to the death of the target cell. b. Fas-FasL Pathway: CTLs express Fas ligand (FasL) on their surface, which can bind to Fas receptors on target cells. This interaction triggers apoptosis in the target cell. c. Secretion of Cytokines: CTLs produce cytokines such as interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), and interleukin-2 (IL-2), which promote inflammation, enhance immune responses, and recruit other immune cells to the site of infection or inflammation.
- Memory Response: Following the clearance of infection, a subset of activated CTLs differentiates into memory CTLs. Memory CTLs provide long-lasting immunity against previously encountered pathogens or antigens. Upon re-exposure to the same antigen, memory CTLs can mount a faster and more robust immune response, leading to rapid elimination of the pathogen or infected cells.
- Regulation and Self-Tolerance: CTL responses are tightly regulated to prevent excessive immune reactions and damage to healthy tissues. Regulatory T cells (Tregs) play a role in suppressing CTL activity and maintaining immune homeostasis. Additionally, CTLs undergo processes of self-tolerance during development in the thymus, ensuring that they do not attack the body's own cells.
- Role in Diseases and Immunotherapy: CTLs are crucial in combating viral infections, controlling intracellular pathogens, and eliminating cancer cells. However, dysregulation or dysfunction of CTLs can contribute to immune-related disorders, chronic infections, and cancers. Immunotherapies such as adoptive cell transfer (ACT) and immune checkpoint inhibitors harness the power of CTLs to enhance immune responses against cancer.
In summary, CTLs are specialized immune cells that recognize and eliminate infected or abnormal cells through cell-mediated cytotoxicity. Their activation, effector functions, memory response, and regulatory mechanisms collectively contribute to immune defense, immune regulation, and the control of diseases. Understanding CTL biology and harnessing their capabilities are essential for developing effective immunotherapies and combating various infections and cancers.
Molecules Associated with the Activity and Function of CTLs
There are many molecules that are closely related to the activity and function of CTLs, including cell surface markers (e.g., CD45, CD28, CD27, CCR7), apoptotic factors (e.g., FASLG), cytotoxins (e.g., GZMA, Granzyme B, GZMH, GZMK, GZMM, PRF1), cytokines (e.g., Ifng, TNFB, TNFa). These molecules help CTLs recognize and destroy infected cells or tumor cells and activate the immune response.
- CD45, also known as leukocyte common antigen (LCA), is a cell surface marker present on various immune cells, including CTLs. CD45 regulates cell signaling through its phosphatase activity and helps CTLs in recognition and activation.
- CD28 is a co-stimulatory molecule present on the surface of CTLs. It binds to CD80 (B7-1) and CD86 (B7-2) on antigen-presenting cells, providing a second signal to activate CTLs and enhance their effector functions.
- CD27 is a co-stimulatory molecule also present on the surface of CTLs. It promotes the survival, expansion, and formation of memory responses in CTLs by binding to its ligand CD70.
- CCR7 is a chemokine receptor present on the surface of CTLs. It helps CTLs to locate and migrate within lymphoid tissues, participating in the regulation of immune responses and the formation of tissue immune memory.
- FASL (FAS ligand) is a cell apoptosis factor present on the surface of CTLs. When CTLs interact with target cells, FASL binds to the FAS (CD95) receptor, triggering apoptosis in the target cells and assisting CTLs in eliminating infected cells or tumor cells.
- GZMA, Granzyme B, GZMH, GZMK, GZMM, PRF1: These are cytotoxic molecules present within CTLs. They induce target cell apoptosis through different mechanisms. GZMA and Granzyme B are the most common cytotoxic molecules, directly activating caspases in target cells to initiate apoptosis. GZMH, GZMK, and GZMM are other types of cytotoxic molecules with potentially different mechanisms of action. PRF1 is a perforin that forms pores on the target cell membrane, allowing cytotoxic molecules to enter the cell.
- Ifng (interferon-gamma), TNFB (tumor necrosis factor-beta), TNFa (tumor necrosis factor-alpha): These are cytokines produced by CTLs. Ifng, an interferon-gamma, participates in the activation of CTLs, promoting cytotoxicity and immune regulation. TNFB and TNFa are members of the tumor necrosis factor family, involved in CTL-mediated cytotoxicity and immune-inflammatory responses, directly targeting tumor cells and infected cells.
These molecules play crucial roles in the regulation of functions and effector functions in CTLs. They aid in the recognition and elimination of infected cells or tumor cells by CTLs and activate immune responses through cell signaling, cell apoptosis, and cytokine production.
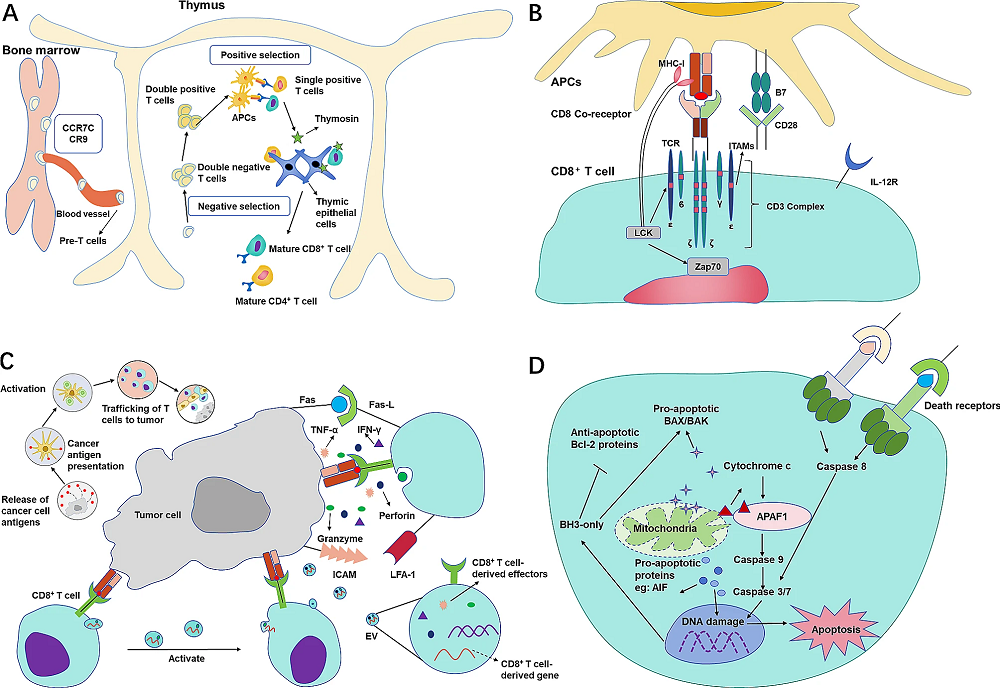 Fig.2 The life cycle of CD8+ T cells. (A) Birth of mature CD8+ T cells. (B) Activation of CD8+ T cells. (C) Anti-tumor effect of CD8+ T cells. (D) The fate of CD8+ T cells. (Chen Y, et al., 2024)
Fig.2 The life cycle of CD8+ T cells. (A) Birth of mature CD8+ T cells. (B) Activation of CD8+ T cells. (C) Anti-tumor effect of CD8+ T cells. (D) The fate of CD8+ T cells. (Chen Y, et al., 2024)
CTLs and Molecules Related to Adaptive Immunity
CTLs are characterized by the expression of the CD8 co-receptor on their surface, which enables them to interact with major histocompatibility complex class I (MHC-I) molecules on target cells. Here's an introduction to the key molecules related to CTLs and adaptive immunity:
- T-cell Receptor (TCR): The TCR is a protein complex expressed on the surface of CTLs and other T cells. It recognizes specific antigens presented by MHC-I molecules on infected or abnormal cells. TCR engagement with antigen-MHC complexes triggers intracellular signaling pathways, leading to CTL activation and effector functions.
- CD8 Co-receptor: The CD8 co-receptor is expressed on the surface of CTLs and enhances the interaction between the TCR and MHC-I molecules. CD8 binding to MHC-I stabilizes the TCR-MHC-I interaction, promoting CTL activation and signaling.
- Major Histocompatibility Complex class I (MHC-I): MHC-I molecules are expressed on the surface of nearly all nucleated cells. They present intracellular peptides derived from endogenous proteins to CTLs. The interaction between the TCR on CTLs and the antigen-MHC-I complex initiates CTL activation and effector functions.
- Co-stimulatory Molecules: Co-stimulatory molecules provide additional signals to CTLs, amplifying their activation and promoting effector functions. Examples of co-stimulatory molecules include CD28, which interacts with CD80 (B7-1) and CD86 (B7-2) on antigen-presenting cells, and CD27, which enhances T cell survival and memory formation.
- Cytokines: Cytokines are small signaling molecules secreted by various immune cells, including CTLs. They regulate immune responses, cell communication, and the differentiation of immune cells. Cytokines produced by CTLs, such as interferon-gamma (IFN-γ) and tumor necrosis factor-alpha (TNF-α), enhance immune activation, promote inflammation, and regulate target cell killing.
- Perforin and Granzymes: CTLs utilize perforin and granzymes to induce target cell death. Perforin is a protein that forms pores in the target cell membrane, allowing the entry of granzymes. Granzymes are serine proteases that initiate apoptosis (programmed cell death) in target cells.
- Fas-Fas Ligand (FasL) Pathway: CTLs can induce apoptosis in target cells through the Fas-FasL pathway. FasL, expressed on the surface of activated CTLs, binds to Fas (CD95) receptors on target cells, triggering cell death.
- Memory T Cells: Upon encountering an antigen, a subset of activated CTLs differentiates into memory T cells. Memory T cells retain the ability to recognize and respond rapidly to previously encountered antigens upon re-infection or re-exposure. They play a crucial role in providing long-lasting immunity.
These molecules and mechanisms are essential for the activation, effector functions, and regulation of CTLs in adaptive immune responses. They collectively contribute to the surveillance and elimination of infected or abnormal cells, helping to maintain immune homeostasis and protect against pathogens and cancer.
The Role of CTLs and Related Molecules in Diseases
- Viral Infections: CTLs are instrumental in controlling viral infections. They recognize virus-infected cells through their T-cell receptors (TCRs) and kill them, limiting viral replication. Molecules such as perforin and granzymes are deployed by CTLs to induce apoptosis in infected cells. Additionally, cytokines produced by CTLs, such as interferon-gamma (IFN-γ), can inhibit viral replication and enhance the immune response against viruses.
- Cancer: CTLs play a crucial role in immunosurveillance and the elimination of cancer cells. Cancer cells can evade immune recognition, but CTLs can recognize specific tumor antigens presented on major histocompatibility complex class I (MHC-I) molecules. CTLs can directly kill cancer cells through the release of cytotoxic molecules like perforin and granzymes. Additionally, cytokines produced by CTLs, such as tumor necrosis factor-alpha (TNF-α), can induce apoptosis in cancer cells and promote anti-tumor immune responses.
- Autoimmune Diseases: In autoimmune diseases, the immune system mistakenly targets and attacks healthy cells and tissues. CTLs can be involved in autoimmune diseases by recognizing self-antigens and initiating immune responses against them. For example, in type 1 diabetes, CTLs target and destroy insulin-producing beta cells in the pancreas. Molecules like CD8, CD28, and cytokines produced by CTLs can contribute to the activation and perpetuation of autoimmune responses.
- Transplant Rejection: CTLs play a significant role in graft rejection following organ transplantation. When a transplanted organ expresses foreign antigens, CTLs can recognize these antigens through their TCRs and initiate an immune response against the graft. CTLs can directly attack and destroy the transplanted cells, leading to organ rejection. Molecules like CD8, perforin, and granzymes are involved in the cytotoxic activity of CTLs during transplant rejection.
- Chronic Inflammatory Diseases: CTLs and their related molecules can contribute to chronic inflammatory diseases, such as rheumatoid arthritis and multiple sclerosis. In these conditions, CTLs can target self-antigens and initiate immune responses against normal tissues. The release of cytokines by CTLs, including IFN-γ and TNF-α, can promote inflammation and tissue damage, contributing to the progression of chronic inflammatory diseases.
Understanding the role of CTLs and related molecules in diseases is crucial for developing targeted therapies and interventions to modulate immune responses and restore immune homeostasis in various pathological conditions.
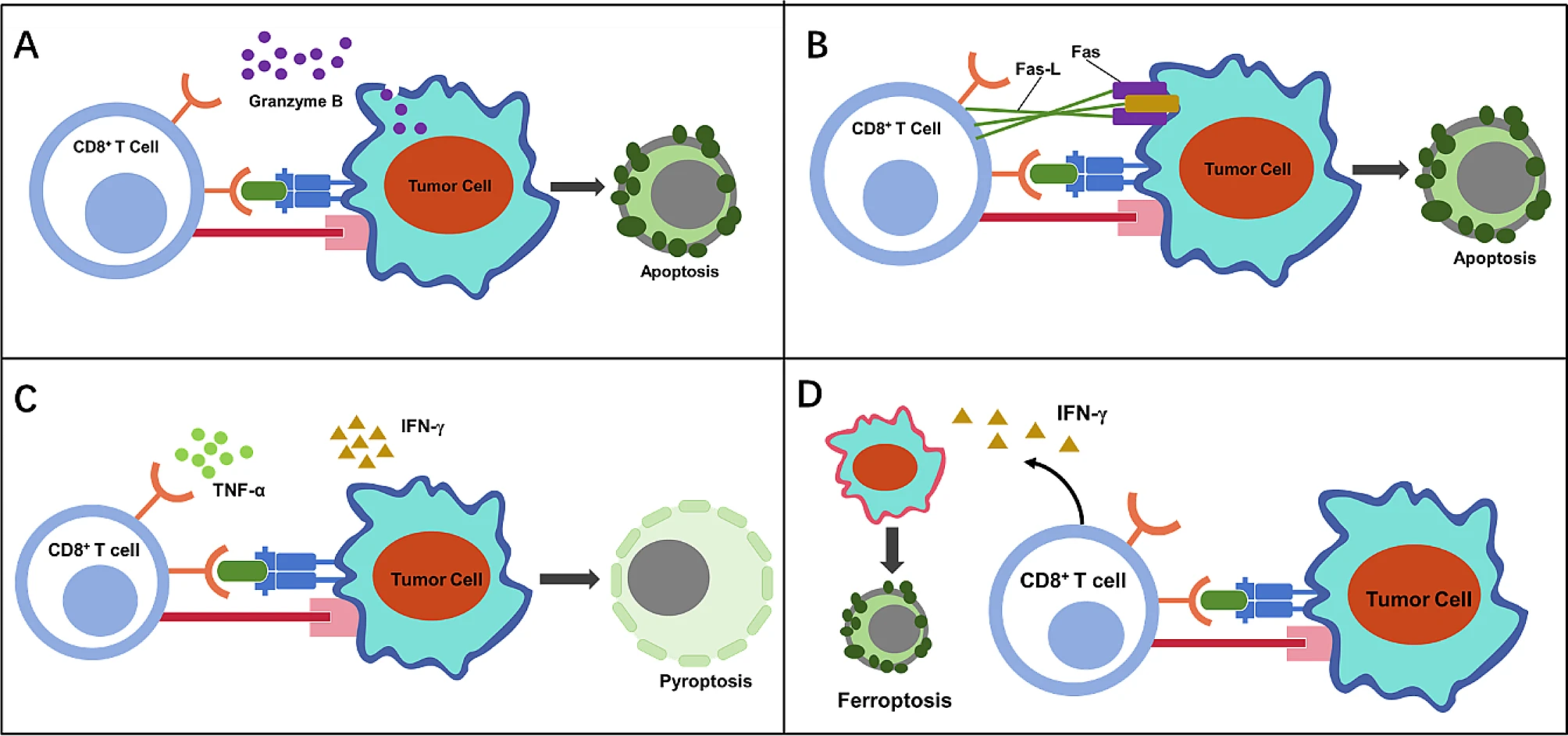 Fig.3 The immunogenic cell death induced by activated CD8+ T cells. (A) CD8+ T cells induce apoptosis of tumor cells through granzyme B release. (B) CD8+ T cells induce apoptosis of tumor cells through death receptor ligand. CD8+ T cells induce (C) pyroptosis (D) or ferroptosis of tumor cells by secreting cytokines. (Chen Y, et al., 2024)
Fig.3 The immunogenic cell death induced by activated CD8+ T cells. (A) CD8+ T cells induce apoptosis of tumor cells through granzyme B release. (B) CD8+ T cells induce apoptosis of tumor cells through death receptor ligand. CD8+ T cells induce (C) pyroptosis (D) or ferroptosis of tumor cells by secreting cytokines. (Chen Y, et al., 2024)
We are dedicated to supporting your research projects and experiments to help you achieve greater success. If you have any questions, needs or cooperation intentions, please feel free to contact us.
Related References
- Weigelin B, den Boer AT, Wagena E, et al. Cytotoxic T cells are able to efficiently eliminate cancer cells by additive cytotoxicity. Nat Commun. 2021;12(1):5217.
- Cenerenti M, Saillard M, Romero P, Jandus C. The Era of Cytotoxic CD4 T Cells. Front Immunol. 2022;13:867189.
- Preglej T, Ellmeier W. CD4+ Cytotoxic T cells - Phenotype, Function and Transcriptional Networks Controlling Their Differentiation Pathways. Immunol Lett. 2022;247:27-42.
- Raskov H, Orhan A, Christensen JP, Gögenur I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br J Cancer. 2021;124(2):359-367.
- Chen Y, Yu D, Qian H, Shi Y, Tao Z. CD8+ T cell-based cancer immunotherapy. J Transl Med. 2024;22(1):394.

