Germ Cell Markers
Related Symbol Search List
Immunology Background
Background
Germ cell markers are specific proteins or antigens that are expressed exclusively in germ cells, which are the precursors of eggs and sperm. These markers are used in research and clinical settings to identify and characterize germ cells, as well as to track their development and differentiation. By studying germ cell markers, scientists can gain insights into reproductive biology, fertility, and various reproductive disorders. Some commonly used germ cell markers include DAZL, OCT4, and SSEA-4. Research on germ cell markers continues to advance our understanding of germ cell biology and holds promise for future applications in assisted reproduction techniques and regenerative medicine.
Roles of Germ Cell Markers
| 1. Developmental Biology | 2. Reproductive Medicine | 3. Stem Cell Research |
| Germ cell markers are essential for understanding gametogenesis—the development process leading to the formation of mature germ cells. By tracing the expression of these markers, researchers can uncover the pathways and signals involved in the differentiation of precursor cells into functional sperm and oocytes. | In assisted reproductive technologies (ART), such as in vitro fertilization (IVF), the identification of germ cell markers can help assess the quality and maturity of gametes. Their expression is critical for evaluating reproductive health, diagnosing infertility issues, and guiding treatment strategies. | Germ cell markers are frequently utilized to characterize pluripotent stem cells, particularly in efforts to derive germ cells from embryonic or induced pluripotent stem cells. This has significant implications for regenerative medicine, offering potential pathways to restore fertility and develop interventions for reproductive disorders. |
Common Germ Cell Markers
Germ cell markers can be classified based on their functions and the specific roles they play during germ cell development and maturation. Commonly used germ cell markers:
- Octamer-binding transcription factor 4 (Oct-4): Maintains pluripotency in embryonic stem cells and is crucial for germ cell development. Expression in primordial germ cells and embryonic germ cells.
- Stella (Dppa3): Essential for germ cell specification and maintenance during early embryonic development. Specific expression in primordial germ cells and germ cell tumors.
- Deleted in Azoospermia-Like (DAZL): Regulates germ cell development and meiosis in both males and females. Expression in primordial germ cells, spermatogonia, and oocytes.
- Vasa Homolog (VASA/DDX4): Plays a key role in germ cell development and gametogenesis. Highly conserved and expressed in germ cells throughout development.
- Dead-End Homolog (DND1): Essential for germ cell migration and maintenance. Expression in primordial germ cells and gonadal germ cells.
- Boule (DDX25): Regulates meiosis and spermatogenesis in male germ cells. Essential for germ cell differentiation in males.
- Nanos: Controls germ cell development and maintenance. Expression in primordial germ cells and germ cell lineage.
- Fragilis (Ifitm3): Involved in germ cell migration and development. Expression in migrating primordial germ cells.
Below are other germ cell markers, organized by prominent categories associated with their functions:
| Classifications | Descriptions |
|---|---|
| RNA-Binding Proteins |
These proteins play crucial roles in the post-transcriptional regulation of gene expression:
|
| Transcription Factors |
These markers are involved in the transcriptional regulation of genes critical for germ cell function:
|
| Gamete-Specific Proteins |
These markers are expressed during the later stages of germ cell differentiation and are specific to mature gametes:
|
| Piwi-Interacting Proteins |
These markers are part of a subclass of proteins that interact with Piwi proteins, crucial for transposon silencing in germ cells:
|
| Accessory Factors and Other Markers |
These markers do not fit neatly into the above categories but are essential for various processes related to germ cell biology:
|
Understanding these germ cell markers is vital for advancing research in reproductive biology, fertility treatments, and understanding germ cell tumors. Their classification based on function helps in elucidating their roles during germ cell development and maturation, contributing to various areas of biomedical research.
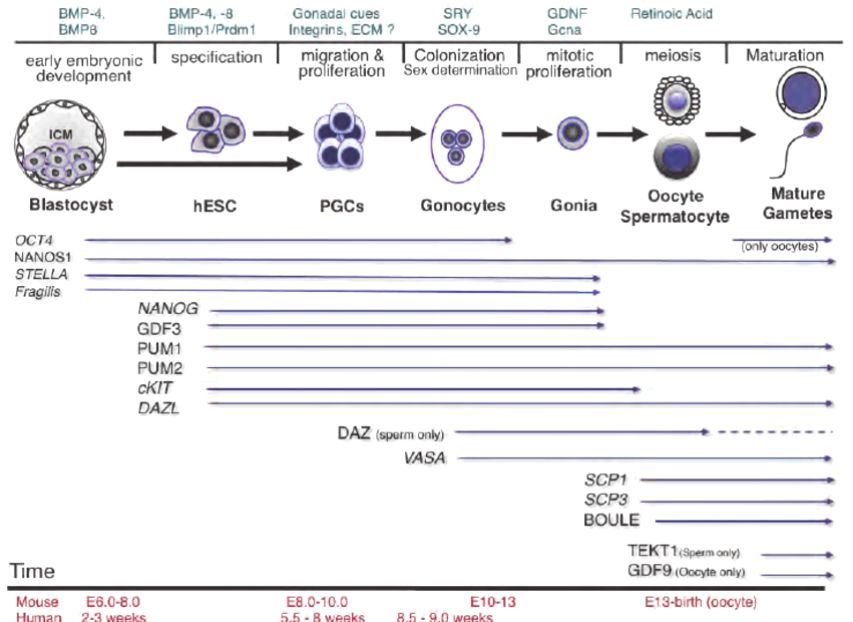 Fig.1 Timeline of germline specification and germ cell marker expression. (Ramathal C, et al., 2011)
Fig.1 Timeline of germline specification and germ cell marker expression. (Ramathal C, et al., 2011)Germ Cell Markers in Various Diseases
Germ cell markers are not only crucial for understanding normal germ cell development but also play significant roles in various diseases, particularly cancers, and reproductive disorders. Certain markers may be re-expressed in malignant germ cell tumors or associated with specific pathologies. Here are some examples of germ cell markers in the context of disease:
Germ Cell Tumors (GCTs)
Germ cell tumors can occur in the gonads or extragonadally. They are categorized as seminomas and non-seminomas. Several germ cell markers are commonly used in diagnosis and treatment monitoring.
- Alpha-fetoprotein (AFP) is one kind of normal reproductive protein in the liver, and it is a spermatocele reproductive cell tumor (NSGCT), and another is a serum specimen of a yolk cyst tumor. In adults, AFP horizontal height may be present in certain cases of testicular cancer.
- Human Chorionic Gonadotropin (hCG) causes reproductive cell production but has a certain type of reproductive cell tumor and secretion, and is particularly known as chorionic gonadotropin (NSGCT). Male hCG is horizontally high, possible presentation of testicular genital cysts, which is likely to occur when AFP is high.
- Placentally derived alkaline phosphatase (PLAP) is a germ cell-specific marker often used in diagnosing seminomas. Increased PLAP levels are often associated with testicular seminomas, aiding in their diagnosis and prognostic evaluation.
Testicular Dysgenesis Syndrome (TDS)
TDS is a condition associated with various reproductive disorders, including infertility, cryptorchidism (undescended testis), and testicular tumors.
- The expression of markers such as Oct4 and Sox2 can be altered in TDS. Abnormalities in the expression of these markers may indicate disrupted germ cell development and maturation, leading to reproductive disorders and increased cancer risk.
Ovarian Cancer
Germ cell markers are also relevant in certain types of ovarian cancer, particularly germ cell tumors.
- VASA (DDX4) is a marker expressed in both normal germ cells and germ cell tumors. VASA can be identified in ovarian germ cell tumors such as dysgerminomas, aiding in diagnosis and classification.
- C-KIT is a part of the receptor tyrosine kinase family, often expressed in embryonic stem cells and germ cells. Abnormal C-KIT signaling has been implicated in some ovarian tumors, suggesting its potential role in tumorigenesis.
Infertility and Reproductive Disorders
Germ cell markers can also provide insights into infertility and other reproductive disorders.
- DAZL (Deleted in Azoospermia-Like) is a gene crucial for spermatogenesis and germ cell development. Mutations in the DAZL gene have been associated with azoospermia (absence of sperm in semen) and other male infertility syndromes.
- Mvh (Mouse Vasa Homologue) is important for germ cell development in various species. Abnormal expression can affect oocyte development and may be linked to female infertility.
Germ cell markers serve as valuable tools in diagnosing and understanding various diseases, particularly cancers and reproductive disorders. Their expression patterns can provide insight into disease mechanisms, prognosis, and potential therapeutic targets. Ongoing research continues to explore the role of these markers in disease, broadening our understanding and guiding clinical practices.
Identification Methods of Germ Cell Markers
Identifying germ cell markers is essential for understanding germ cell development and function. Several methods are employed to identify and characterize these markers. Here are some of the most common identification methods:
1. Molecular Biology Techniques
a. Gene Expression Analysis
RT-PCR is used to measure mRNA levels of specific germ cell markers. It allows for the quantification of gene expression in germ cells.
qPCR is a more sensitive version of PCR that provides quantitative data on gene expression, often used after RT-PCR.
RNA-Seq high-throughput sequencing provides comprehensive data on the transcriptome, allowing researchers to identify novel germ cell markers by comparing germ cells and somatic cells.
b. In Situ Hybridization
This technique involves using labeled probes that hybridize to specific mRNA transcripts in tissue sections. This allows for the visualization of the spatial expression patterns of germ cell markers in intact tissues.
2. Protein Analysis
Western Blotting is used to detect specific proteins associated with germ cells by separating proteins based on size and using antibodies specific to germ cell markers.
Immunofluorescence and immunohistochemistry involve the use of antibodies that are tagged with fluorescent markers (immunofluorescence) or enzymes (immunohistochemistry). They allow for the visualization of the distribution of germ cell markers in cells and tissues under a microscope.
3. Flow Cytometry
Flow cytometry can be used to analyze the expression of surface proteins on germ cells. It allows for the isolation and characterization of germ cell populations based on specific markers.
4. Chromatin Immunoprecipitation (ChIP)
ChIP can be used to study the binding of transcription factors and modify histones associated with germ cell-specific genes, providing insights into regulatory mechanisms controlling germ cell marker expression.
5. Functional Studies
a. Gene Knockout/Knockdown
By knocking out or knocking down specific genes believed to code for germ cell markers (using CRISPR/Cas9 or RNA interference), researchers can assess the effects on germ cell development and confirm the role of these markers.
b. Transgenic Models
Creating transgenic animals (e.g., mice) that express reporter genes under the control of germ cell-specific promoters can help visualize the expression patterns of germ cell markers in vivo.
6. Comparative Studies
Researchers often compare gene expression profiles in germ cells with those in somatic cells using various model organisms (e.g., mice, zebrafish, humans) to identify conserved germ cell markers across species.
7. Bioinformatics Approaches
Analyzing existing genomic and transcriptomic databases can help identify putative germ cell markers based on their expression patterns in known germ cell populations.
Identifying germ cell markers involves a combination of molecular biology, protein analysis, functional studies, and computational approaches. The integration of these methods helps provide a comprehensive understanding of germ cell development, their functions, and their relevance in health and disease, facilitating advancements in reproductive biology and medicine.
Case Study
Case 1: Heeren AM, He N, de Souza AF, et al. On the development of extragonadal and gonadal human germ cells. Biol Open. 2016;5(2):185-194.
Human germ cells have an extragonadal origin and migrate to colonize the gonadal primordia around seven weeks of gestation. During this migration, many germ cells are lost through apoptosis, but some evade this fate, potentially leading to extragonadal germ cell tumors. Investigating the presence of ectopic germ cells in human adrenals, researchers discovered the existence of germ cells expressing DDX4 and/or POU5F1 in male and female human adrenals during the first and second trimesters. Unlike observations in mice, where adrenal and ovarian germ cells appear to enter meiosis simultaneously, human adrenal germ cells did not exhibit meiotic entry until week 22. In contrast, ovarian germ cells at week 22 displayed asynchronous meiotic initiation. Notably, immature POU5F1+ germ cells in early human ovaries retained expression of the neural crest marker TUBB3, reminiscent of their migratory phase. These findings underscore species-specific variations in early gametogenesis between mice and humans and reveal the presence of ectopic germ cell populations in human adrenals during development.
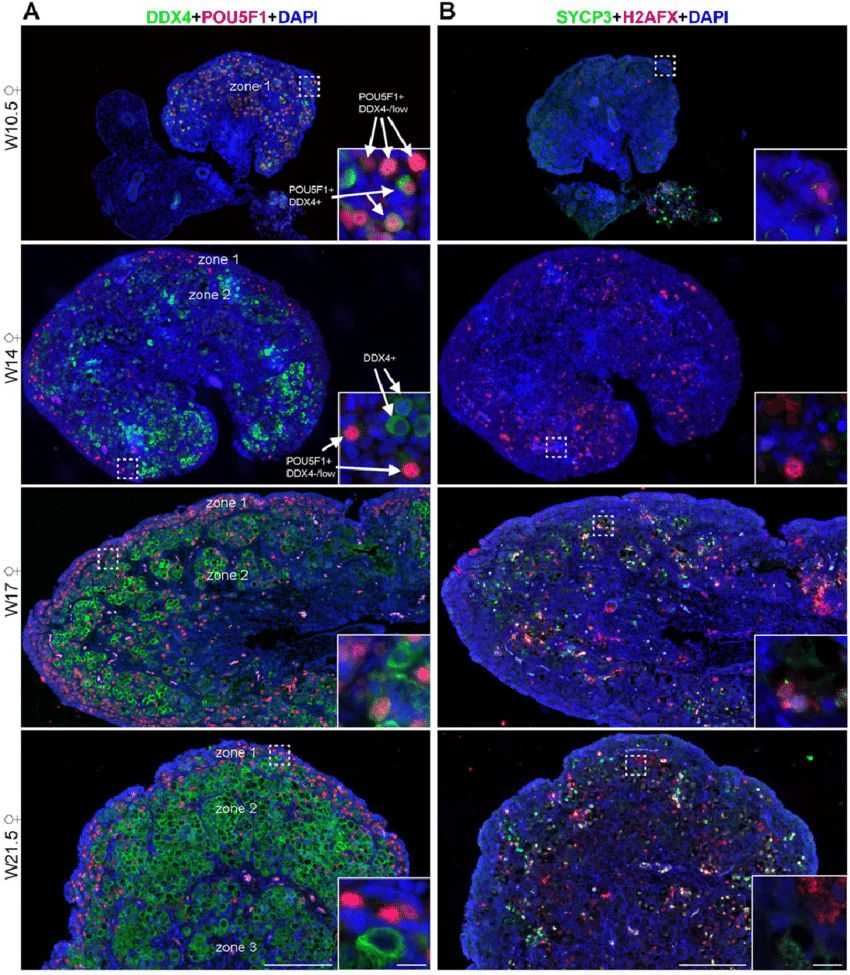 Fig.1 Dynamic expression of germ cell markers in human fetal ovaries.
Fig.1 Dynamic expression of germ cell markers in human fetal ovaries.Case 2: Zhao, J., Lu, P., Wan, C., et al. Cell-fate transition and determination analysis of mouse male germ cells throughout development. Nat Commun 12, 6839 (2021).
In mammalian male germ cell development, a sequential progression of cell fate changes occurs, yet a detailed understanding of the complete developmental trajectory remains elusive. Through an analysis of a detailed transcriptome atlas encompassing 11,598 cells across 28 crucial time points, researchers have illustrated that the shift from mitotic to post-mitotic primordial germ cells involves significant transcriptome reconfiguration and the emergence of a transitional cell state. The Notch signaling pathway plays a pivotal role in initiating mitotic arrest and preserving the identities of male germ cells. Experimentally induced HELQ ablation results in developmental stalling and aberrant transcriptome restructuring in male germ cells, emphasizing the importance of regulating the cell cycle for proper cell fate transitions. Furthermore, a comparative analysis between humans and mice uncovers potential regulators whose deficiencies may lead to male infertility in humans through the regulation of mitotic arrest. This study offers a detailed and thorough transcriptome map of the male germline cycle, enhancing our understanding of the mechanisms governing cell fate transitions and determinations during male germ cell development.
In this study, researchers further defined 9 subgroups from SPC and RS based on dynamic expression of stage PGC-specific markers. The continuity of all germ cell types matched the progression of the intensively sampled time points. In addition, four major somatic cell clusters were annotated as progenitors, peritubular myeloid and interstitial progenitors (PMC and IP), mesenchymal cells, and sertoli cells, based on previously identified cell type markers.
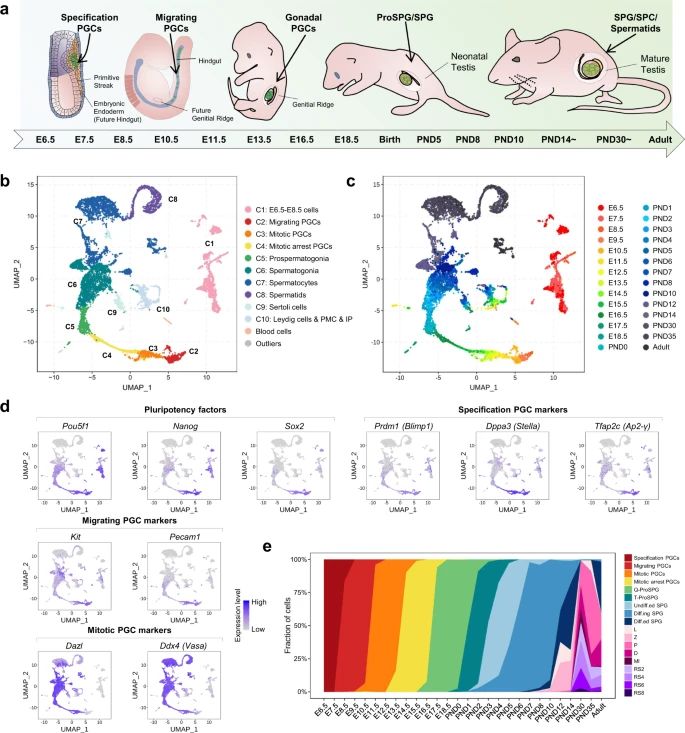 Fig.2 A single-cell map of mouse male germ cell development.
Fig.2 A single-cell map of mouse male germ cell development.Case 3: Hwang YS, Suzuki S, Seita Y, et al. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat Commun. 2020;11(1):5656.
The establishment of spermatogonia during both fetal development and postnatal stages is crucial for the continuous production of sperm and male fertility. In this study, a method is devised for the in vitro reconstruction of human prospermatogonial specification. Human primordial germ cell (PGC)-like cells, derived from human induced pluripotent stem cells, are differentiated into M-prospermatogonia-like cells and T1 prospermatogonia-like cells (T1LCs) through extended culture in xenogeneic reconstituted testes. By employing single-cell RNA sequencing, the lineage trajectory leading to T1LCs is mapped out, revealing that these cells closely resemble human T1-prospermatogonia in vivo. They exhibit gene expression patterns associated with spermatogenesis and reduced proliferation, characteristic of quiescent T1 prospermatogonia. Significantly, this system allows for the observation of the dynamic and stage-specific regulation of transposable elements during human prospermatogonial specification. These results open up new possibilities for studying and replicating human male germline development in vitro.
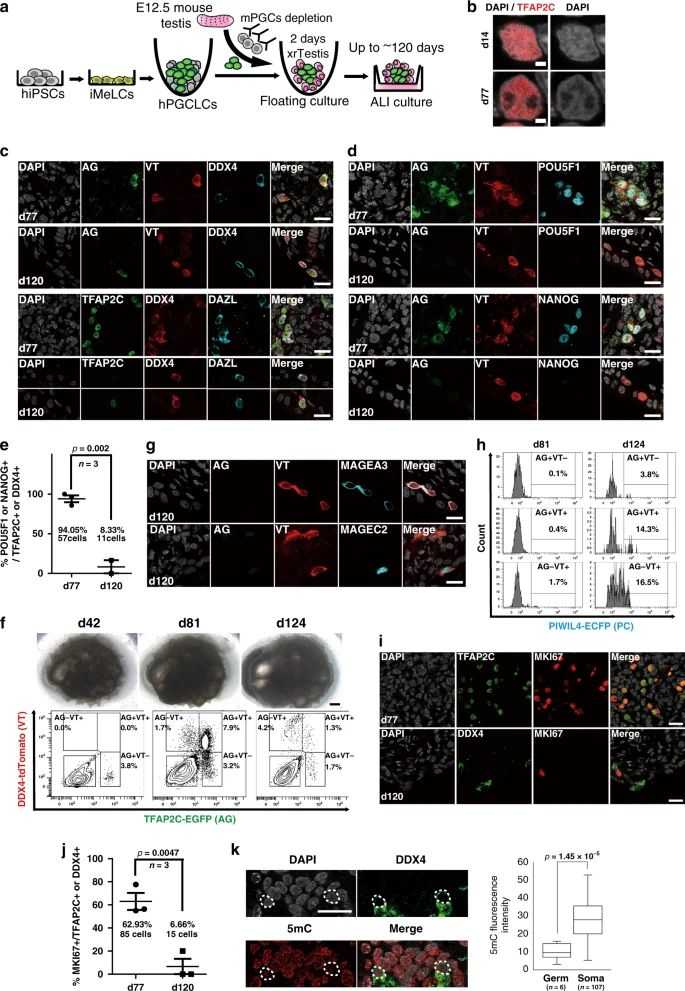 Fig.3 Establishment of xenogeneic reconstituted testis (xrTestis) and generation of human T1LCs.
Fig.3 Establishment of xenogeneic reconstituted testis (xrTestis) and generation of human T1LCs.Related References
- Ramathal C, Pera R R, Turek P. Embryonic stem cells and the germ cell lineage //Embryonic stem cells-basic biology to bioengineering. IntechOpen, 2011.
- Mathan, Zaib G, Jin K, Zuo Q, Habib M, Zhang Y, Li B. Formation, application, and significance of chicken primordial germ cells: A Review. Animals. 2023; 13(6):1096.

