ADP-ribosyltransferases (ARTs)
Related Symbol Search List
Immunology Background
Background
ADP-ribosyltransferases (ARTs) are a family of enzymes that catalyze the transfer of ADP-ribose moieties from NAD+ (nicotinamide adenine dinucleotide) to target proteins, leading to the addition of ADP-ribose modifications. These modifications play important roles in various cellular processes, including DNA repair, chromatin remodeling, transcriptional regulation, signal transduction, and cellular stress responses.
ARTs typically possess a conserved catalytic domain known as the ART domain. This domain contains the catalytic site responsible for the transfer of ADP-ribose. In the 1980s and 1990s, the name poly(ADP-ribose) synthetase (PARS) as well as ADP-ribosyltransferase (ADPRT) were used, but then gradually replaced by PARS. Later, PARPx and TNKSx were used as names for ARTD family members. In eukaryotes, ARTD summarizes the intracellular ART family with PARP and TNKS as the names for individual proteins that possess a diphtheria toxin-like version of the ART fold, while ARTC describes the family of extracellular enzymes with a cholera toxin-like version of the ART fold. ARTs typically possess a conserved catalytic domain known as the ART domain. This domain contains the catalytic site responsible for the transfer of ADP-ribose.
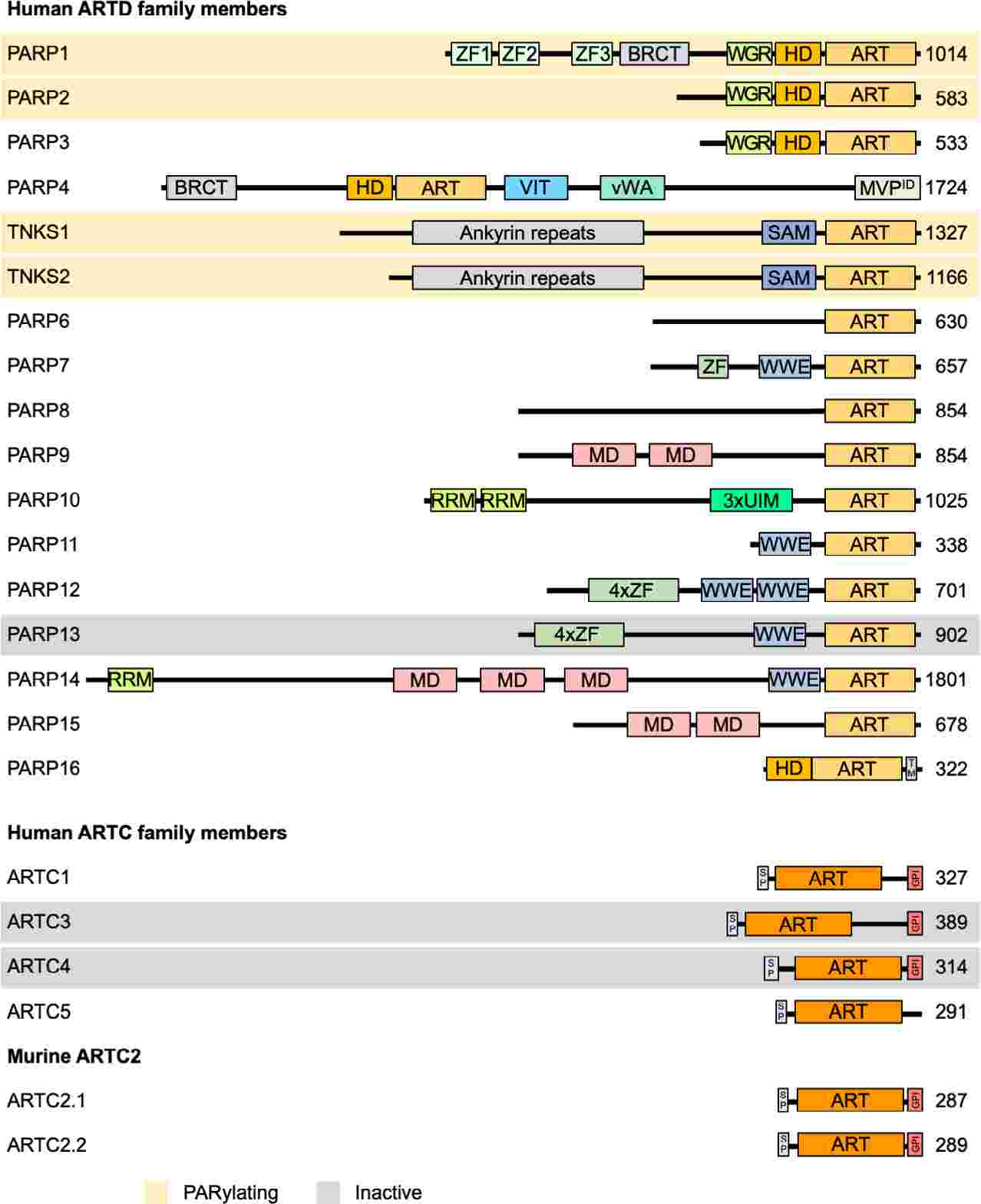 Fig.1 Domain architecture of ARTD and ARTC family members. (Lüscher B, et al., 2022)
Fig.1 Domain architecture of ARTD and ARTC family members. (Lüscher B, et al., 2022)Members of the ARTs Family
| Members | Details |
|---|---|
| PARP1 | PARP1 (Poly-ADP-ribose polymerase 1) is a well-studied member of the ART family and plays a crucial role in DNA repair processes, particularly in base excision repair. It is involved in the detection and repair of DNA single-strand breaks by catalyzing the addition of poly-ADP-ribose chains to target proteins. |
| PARP2 | PARP2 shares structural and functional similarities with PARP1 and is also involved in DNA repair. It participates in base excision repair and contributes to the maintenance of genome stability. |
| ARTD1 | ARTD1 (ADP-ribosyltransferase diphtheria toxin-like 1, also known as PARP3) is a mono-ADP-ribosyltransferase that plays a role in DNA repair and genome maintenance. It is involved in the repair of DNA double-strand breaks and interacts with other DNA repair proteins. |
| ARTD2 | ARTD2 (also known as PARP2) is a mono-ADP-ribosyltransferase that has been implicated in various cellular processes, including transcriptional regulation, DNA repair, and cell death pathways. It interacts with transcription factors and chromatin modifiers to modulate gene expression. |
| ARTD5 | ARTD5 (also known as ART5) is a mono-ADP-ribosyltransferase involved in the regulation of cell proliferation and survival. It interacts with p53, a tumor suppressor protein, and modulates its activity. |
| ARTC1 | ARTC1 (ADP-ribosyltransferase C1, also known as ART1) is a cell surface enzyme that catalyzes the ADP-ribosylation of target proteins on the extracellular side of the plasma membrane. It plays a role in immune responses and has been implicated in the regulation of T-cell activation and function. |
These are just a few examples of the ADP-ribosyltransferases within the ART family. There are additional members of this enzyme family with distinct functions and substrate specificities, contributing to the diverse roles of ADP-ribosylation in cellular processes.
Mechanism of Action for ARTs
ARTs catalyze the transfer of ADP-ribose moieties from NAD+ to target proteins, thereby modifying the target proteins' structure and function. The mechanism of action for ARTs involves several steps:
- Substrate Recognition: ARTs recognize specific target proteins or amino acid residues within proteins as substrates. The substrate specificity varies among different ARTs and is determined by the enzyme's catalytic domain and other structural features.
- NAD+ Binding: ARTs bind to NAD+, which serves as the donor molecule for the ADP-ribose group. NAD+ is a coenzyme involved in various cellular processes and is abundant in cells.
- ADP-ribose Transfer: The catalytic domain of ARTs contains a conserved catalytic site. In this site, the enzyme cleaves the glycosidic bond between the nicotinamide and ribose moieties of NAD+, generating nicotinamide and an ADP-ribose intermediate.
- ADP-ribose Transfer to Substrate: The ADP-ribose intermediate is then transferred to the target protein. The specific amino acid residues within the target protein, such as aspartic acid, glutamic acid, or arginine, serve as acceptor sites for the ADP-ribose group.
- ADP-ribosylation of Substrate: The ADP-ribose group is covalently attached to the acceptor amino acid residue within the target protein, forming an ADP-ribosylated protein. This modification can occur as a single ADP-ribose unit (mono-ADP-ribosylation) or as multiple ADP-ribose units, forming poly-ADP-ribose chains (poly-ADP-ribosylation), depending on the specific ART involved.
Regulation of ART Activity and Function
The activity and function of ARTs can be regulated through various mechanisms:
| Activity and Function | Details |
|---|---|
| Post-translational Modifications | The amino acids being modified during protein ADP-ribosylation are various, the attachment may occur on carboxylic groups, such as on Aspartate (D) and Glutamate (E), on amino or guanidino groups, such as in Lysine (K) and Arginine (R), thiol group in Cysteine (C) (non-enzymatic attachment), and on hydroxyl groups as in Serine (S). Therefore, various and different post-translational modifications may modify the same acceptor amino acid, precluding its modification by another type of enzyme, or may impede the access to a neighbor site by hindrance, or by repulsing charges. |
| Protein-Protein Interactions | ARTs can interact with other proteins, including regulators, co-factors, or target proteins, which can influence their activity. These interactions may promote or inhibit the enzymatic function of ARTs or provide specificity for substrate recognition. |
| Substrate Availability | The availability of target proteins or specific amino acid residues within substrates can affect the activity of ARTs. Changes in cellular conditions or signaling pathways can modulate the expression, localization, or accessibility of target proteins, thereby influencing the substrate pool for ARTs. |
| Regulatory Molecules | Certain molecules, such as metabolites or signaling molecules, can directly or indirectly regulate ART activity. These molecules can modulate the catalytic activity, substrate specificity, or binding affinity of ARTs, thereby impacting their function. |
Overall, the activity and function of ARTs are tightly regulated to ensure precise control of ADP-ribosylation processes in cells. The interplay between ARTs, their substrates, and regulatory factors contributes to the dynamic and context-dependent nature of ADP-ribosylation in cellular processes.
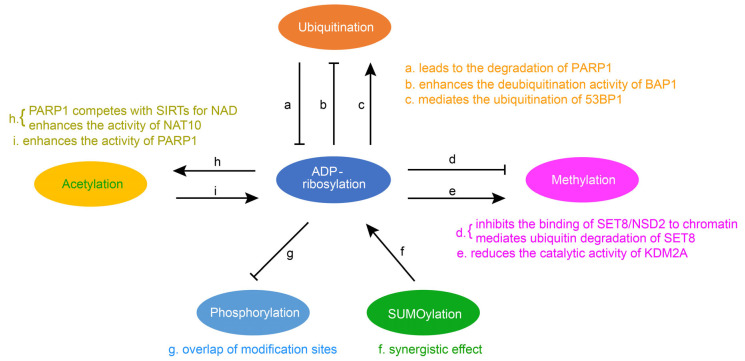 Fig.2 Crosstalk of ADP-ribosylation with other protein post-translational modifications. (Li Z, et al., 2023)
Fig.2 Crosstalk of ADP-ribosylation with other protein post-translational modifications. (Li Z, et al., 2023)Biological Functions of the ARTs Family
ARTs play crucial roles in various biological functions within cells. Here are some key roles of ARTs in cellular processes:
Apoptosis
ARTs are involved in regulating apoptotic pathways, which are essential for programmed cell death. ADP-ribosylation by ARTs can modulate the activity and function of proteins involved in apoptotic signaling. For example, ARTs can ADP-ribosylate apoptotic regulators like p53, Bcl-2 family members, and caspases, influencing their stability, interactions, and enzymatic activity. These modifications can promote or inhibit apoptotic signaling, contributing to the regulation of cell survival and programmed cell death processes.
DNA Repair
ARTs, particularly poly-ADP-ribose polymerases (PARPs), are critically involved in DNA repair processes. PARPs are recruited to sites of DNA damage and catalyze the addition of poly-ADP-ribose chains to target proteins, including histones and DNA repair factors. This modification facilitates the assembly and recruitment of DNA repair complexes, signaling DNA damage, and promoting efficient repair of DNA lesions. PARPs are particularly important in base excision repair and DNA strand break repair pathways, ensuring genome stability and preventing the accumulation of DNA damage.
Chromatin Remodeling and Transcriptional Regulation
ARTs influence chromatin structure and transcriptional regulation through ADP-ribosylation of histones, transcription factors, and chromatin modifiers. ADP-ribosylation can modulate chromatin accessibility, histone modifications, and interactions between transcription factors and DNA. By altering these processes, ART-mediated ADP-ribosylation can regulate gene expression patterns, transcriptional activation or repression, and cellular responses to various stimuli.
Signal Transduction
ARTs participate in signal transduction pathways by modifying key signaling molecules. ADP-ribosylation can impact the activity, stability, subcellular localization, or interactions of signaling proteins, thereby regulating signal transduction cascades. For example, ARTs can modify GTPases, kinases, and other signaling molecules involved in cell growth, differentiation, immunity, and stress responses. ADP-ribosylation can modulate protein-protein interactions, enzymatic activities, and downstream signaling events, influencing cellular responses to extracellular cues.
Immune Responses
ARTs, such as ARTC1, play roles in immune responses and immune cell functions. ARTC1, a cell surface ADP-ribosyltransferase, is involved in regulating T-cell activation and function. ADP-ribosylation by ARTC1 can impact the function and activation of immune receptors and signaling molecules involved in immune cell signaling and responses.
These are just a few examples of the diverse roles of ARTs in cellular biology. ARTs and ADP-ribosylation processes have far-reaching effects on cell physiology, including cell survival, genome integrity, gene expression, and cellular responses to various stimuli. The precise regulation of ART activity and substrate specificity ensures the proper functioning of these biological processes.
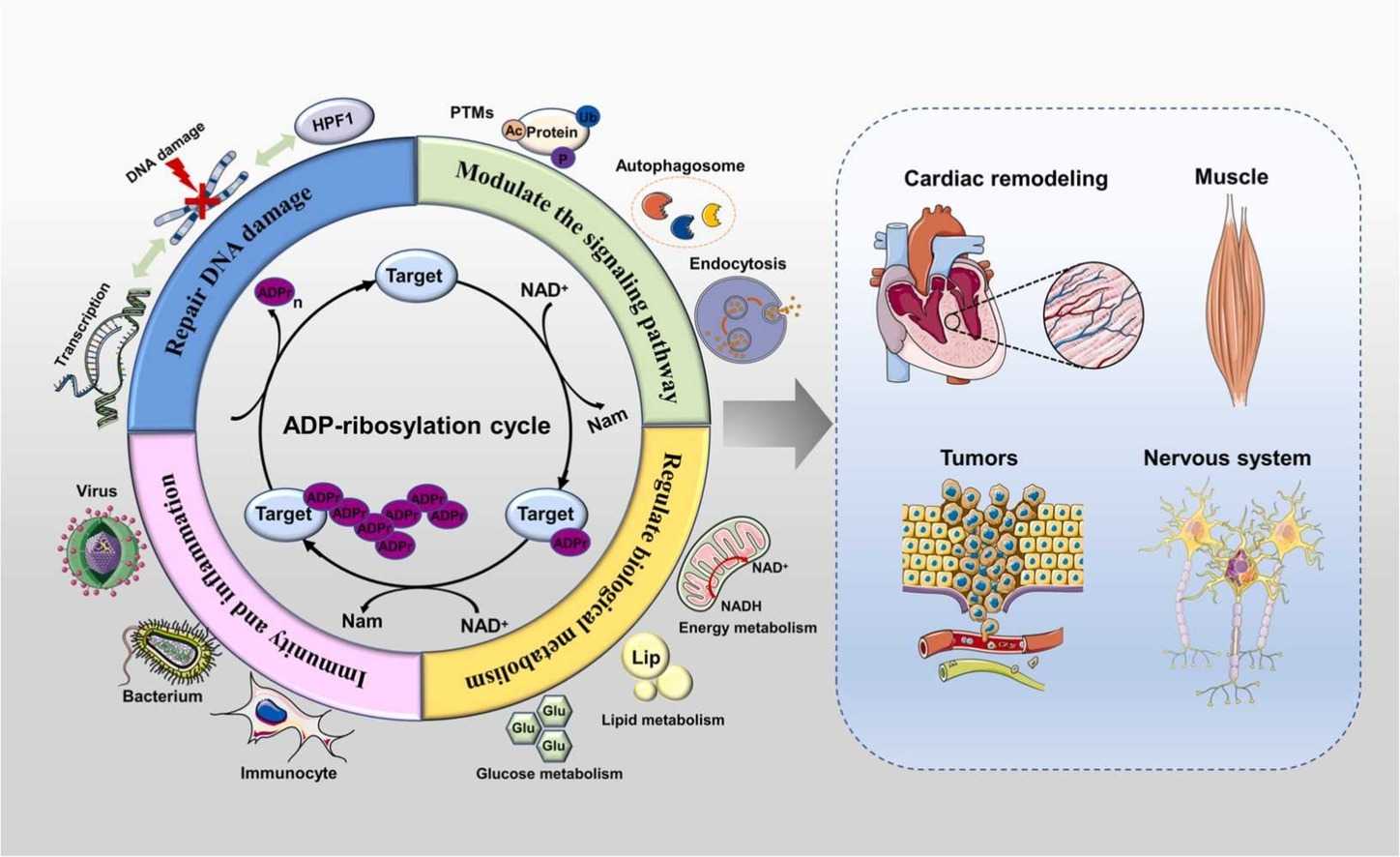 Fig.3 ADP-ribosylation (ADPr) modification is involved in signaling pathways, DNA damage repair, metabolism, immunity, and inflammation. (Liu YT, et al., 2024)
Fig.3 ADP-ribosylation (ADPr) modification is involved in signaling pathways, DNA damage repair, metabolism, immunity, and inflammation. (Liu YT, et al., 2024)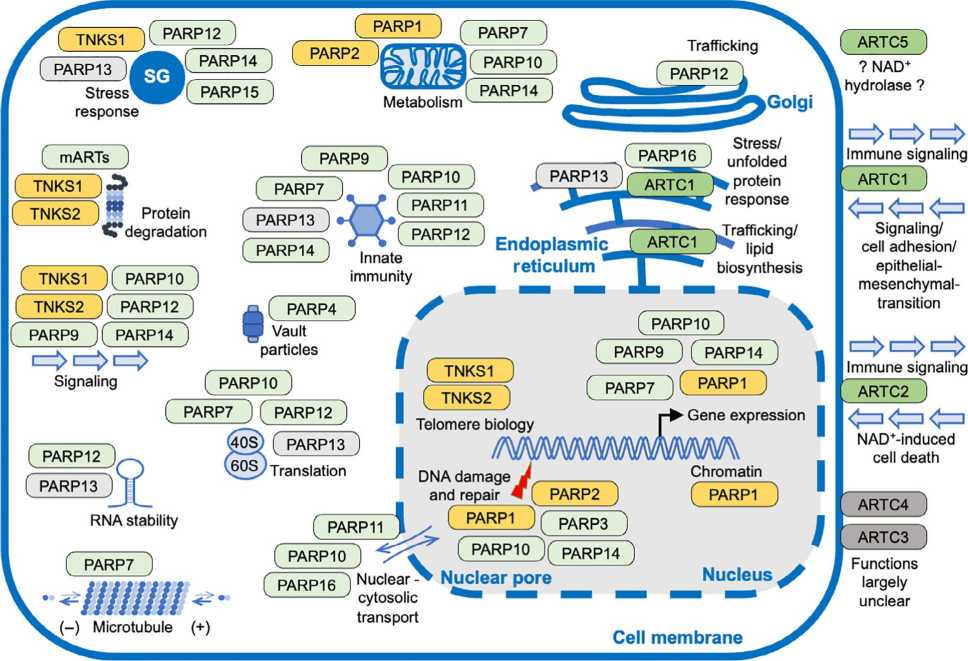 Fig.4 Summary of the functions of mammalian ART family members. (Lüscher B, et al., 2022)
Fig.4 Summary of the functions of mammalian ART family members. (Lüscher B, et al., 2022)Case Study
Case 1: Li L, Shi Y, Li S, et al. ADP-ribosyltransferase PARP11 suppresses Zika virus in synergy with PARP12. Cell Biosci. 2021;11(1):116.
Zika virus (ZIKV) infection is a global health threat associated with microcephaly and other neurological complications. The study aimed to investigate the anti-ZIKV mechanism of PARP11. By generating PARP11-knockout and PARP11-overexpressing A549 cell lines, the researchers evaluated the impact of PARP11 on ZIKV replication. The results demonstrated that ZIKV replication was suppressed in PARP11-overexpressing cells and enhanced in PARP11-knockout cells. The growth kinetics of ZIKV also supported these findings. Therefore, PARP11 is identified as an interferon-stimulated gene (ISG) with anti-ZIKV activity, suggesting its role in innate immune defense against ZIKV replication.
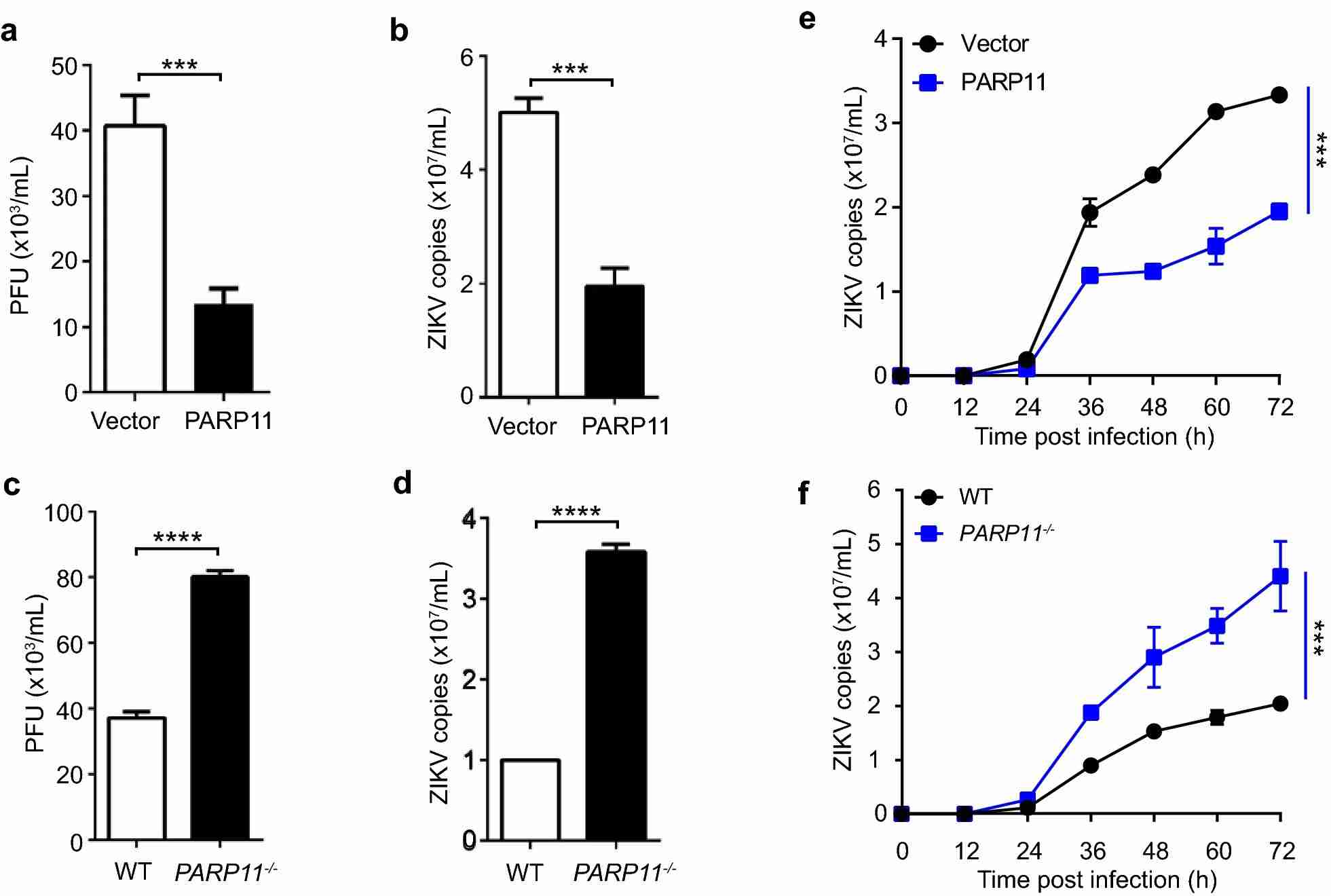 Fig.1 PARP11 suppresses ZIKV replication in vitro.
Fig.1 PARP11 suppresses ZIKV replication in vitro.Case 2: Mayo E, Fabrizio G, Scarpa ES, Stilla A, et al. ARTD10/PARP10 induces ADP-ribosylation of GAPDH and recruits GAPDH into cytosolic membrane-free cell bodies when overexpressed in mammalian cells. Challenges. 2018; 9(1):22.
The study focuses on the interaction between the glycolytic enzyme GAPDH and the enzyme ARTD10/PARP10, a member of the ADP-ribosyltransferase family. The researchers found that GAPDH is an interactor and a novel target for ARTD10/PARP10. They observed the co-localization of GAPDH and ARTD10/PARP10 in membrane-free cytosolic bodies. By using immunofluorescence microscopy and inhibitory compounds, they demonstrated that the presence of GAPDH in these cytosolic bodies depends on the catalytic activity of ARTD10/PARP10. Additionally, when the catalytic domain of ARTD10/PARP10 alone was overexpressed, GAPDH was recruited into stress granules.
The study investigated whether GAPDH can be ADP-ribosylated by ARTD10. Pull-down assays and in vitro ADP-ribosylation assays confirmed that ARTD10 can specifically ADP-ribosylate GAPDH. These findings highlight ARTD10/PARP10 as the enzyme responsible for modifying and recruiting GAPDH into cytosolic structures.
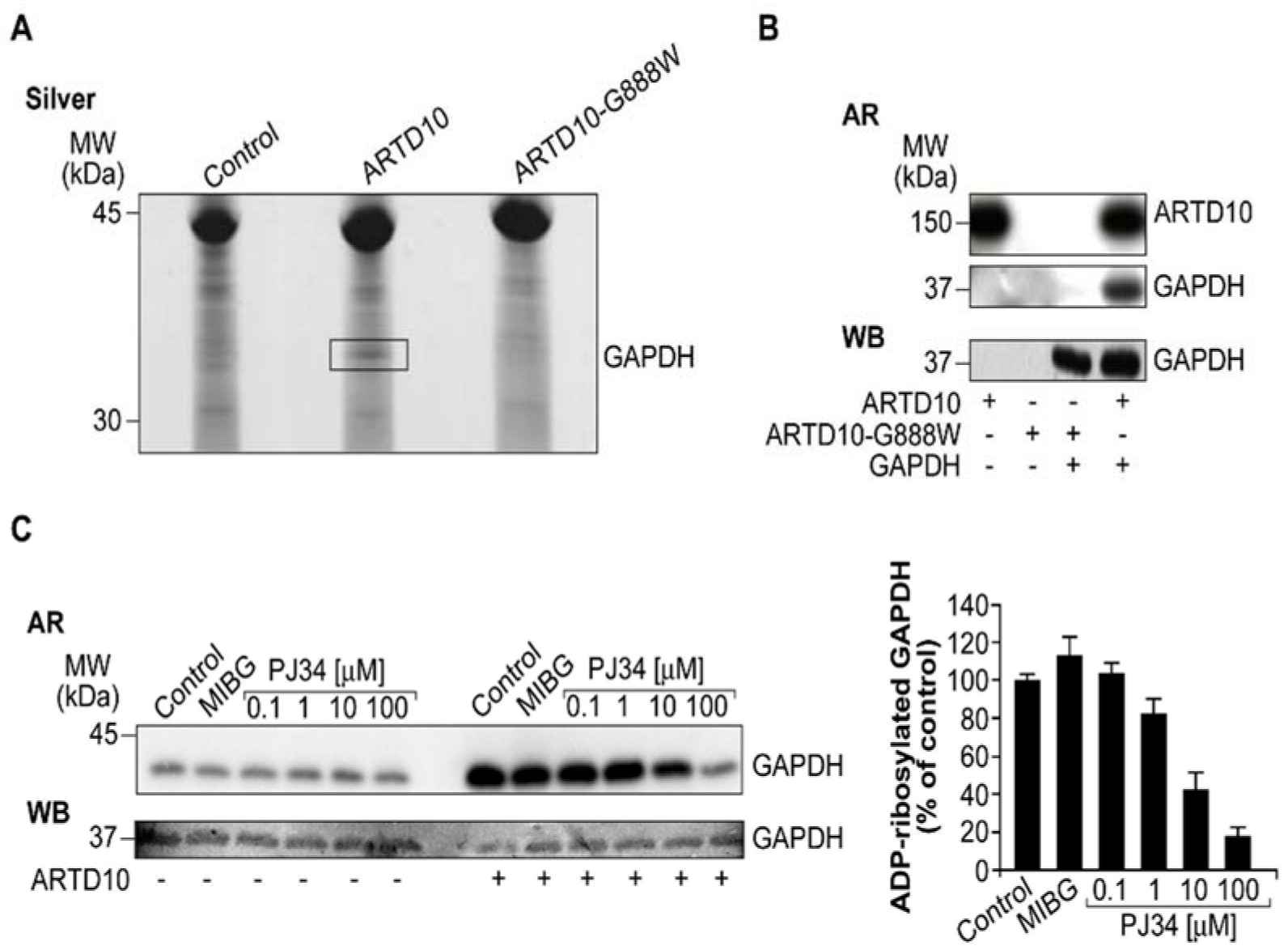 Fig.2 ARTD10/PARP10 ADP-ribosylates GAPDH.
Fig.2 ARTD10/PARP10 ADP-ribosylates GAPDH.Related References
- Lüscher B, Ahel I, Altmeyer M, et al. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 2022;289(23):7399-7410.
- Li Z, Luo A, Xie B. The complex network of ADP-ribosylation and DNA repair: emerging insights and implications for cancer therapy. Int J Mol Sci. 2023;24(19):15028.
- Liu YT, Che Y, Qiu HL, et al. ADP-ribosylation: An emerging direction for disease treatment. Ageing Res Rev. 2024;94:102176.

