Phosphatases in the Akt Pathway
Related Symbol Search List
- ACP1
- Acid Phosphatase
- CALM1
- CDC25A
- DUSP1
- DUSP6
- EPM2A
- FKBP12
- FKBP5
- PPIG
- PPP1CA
- PPP1R1B
- PPP3CA
- PPP3R1
- PPP3R2
- PTEN
- PTP4A2
- PTP4A3
- PTP1B
- PTPN11
- PTPN2
- SIRPA
- CD45
- PTPRJ
Immunology Background
About Phosphatases in the Akt Pathway
The Akt pathway, also known as the PI3K/Akt pathway, is a critical signaling pathway involved in cell growth, survival, metabolism, and other cellular processes. Phosphatases play a crucial role in regulating the activation and signaling of Akt, a central kinase within this pathway.
Phosphatases are enzymes responsible for removing phosphate groups from proteins, counteracting the actions of kinases that add phosphate groups during phosphorylation. In the Akt pathway, protein phosphatases are involved in dephosphorylating Akt, thereby modulating its activity and downstream signaling.
One of the key phosphatases involved in the Akt pathway is Protein Phosphatase 2A (PP2A). PP2A is a serine/threonine phosphatase that regulates Akt by dephosphorylating specific residues critical for its activation. PP2A is a heterotrimeric complex consisting of a catalytic C subunit, a structural A subunit, and a regulatory B subunit. The regulatory subunit confers substrate specificity and subcellular localization to the PP2A complex.
The Akt pathway is activated in response to extracellular signals, such as growth factors, hormones, and cytokines. Activation of the pathway begins with the binding of these ligands to cell surface receptors, leading to the activation of Phosphatidylinositol 3-Kinase (PI3K). Activated PI3K phosphorylates phosphatidylinositol 4,5-bisphosphate (PIP2) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3) at the plasma membrane. PIP3 acts as a second messenger and recruits Akt to the plasma membrane through its pleckstrin homology (PH) domain.
Once localized to the plasma membrane, Akt undergoes phosphorylation at two critical sites: Threonine 308 (Thr308) and Serine 473 (Ser473). Phosphorylation at Thr308 is mediated by Phosphoinositide-Dependent Kinase 1 (PDK1), while phosphorylation at Ser473 is mediated by Mammalian Target of Rapamycin Complex 2 (mTORC2). These phosphorylation events are crucial for the full activation of Akt.
However, the activity of Akt is regulated by protein phosphatases, including PP2A, which dephosphorylate Akt and counteract its activation. PP2A specifically targets the phosphorylated Thr308 and Ser473 residues on Akt, leading to their dephosphorylation and subsequent inactivation. This dephosphorylation process negatively regulates Akt activity and downstream signaling.
The balance between kinase-mediated phosphorylation and phosphatase-mediated dephosphorylation is critical for the precise control of Akt signaling. Dysregulation of this balance can result in aberrant Akt activation, which is associated with various diseases, including cancer, neurodegenerative disorders, and metabolic disorders.
In summary, phosphatases play a crucial role in the Akt pathway by dephosphorylating and inactivating Akt. They act as negative regulators that counterbalance the activating phosphorylation events mediated by kinases, ensuring proper control of Akt signaling. Understanding the interplay between kinases and phosphatases in the Akt pathway is vital for unraveling the regulatory mechanisms and their implications in various physiological and pathological processes.
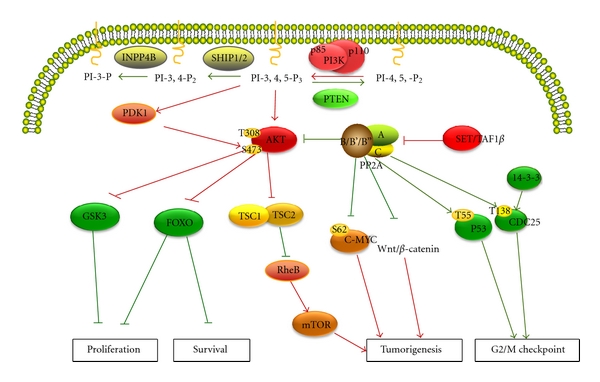 Fig.1 The primary phosphatases function as tumor suppressors and their signaling pathways. (Zhang Q, et al., 2012)
Fig.1 The primary phosphatases function as tumor suppressors and their signaling pathways. (Zhang Q, et al., 2012)
Biological Functions of Phosphatases in the Akt Pathway
Negative Regulation of Akt Activation: Phosphatases, such as PP2A, play a critical role in negatively regulating Akt activation. After growth factor stimulation, Akt is phosphorylated at Thr308 by Phosphoinositide-Dependent Kinase 1 (PDK1) and at Ser473 by Mammalian Target of Rapamycin Complex 2 (mTORC2). Phosphatases, including PP2A, dephosphorylate these phosphorylated residues on Akt, leading to its deactivation. This dephosphorylation inhibits Akt activity and prevents its downstream signaling, thereby maintaining a proper balance in the Akt pathway.
Control of Cell Growth and Survival: Phosphatases in the Akt pathway play crucial roles in regulating cell growth and survival. Akt is a key mediator of cell survival signals and promotes cell growth by stimulating protein synthesis and inhibiting apoptosis. Phosphatases, by dephosphorylating Akt, counteract its pro-survival and growth-promoting actions, thereby regulating cell fate decisions.
Regulation of Glycogen Metabolism: Akt plays a significant role in regulating glycogen metabolism by promoting glycogen synthesis and inhibiting glycogen breakdown. Phosphatases, including PP2A, contribute to the regulation of glycogen metabolism by dephosphorylating and inactivating Akt. This dephosphorylation event limits Akt's ability to stimulate glycogen synthesis, ensuring proper glycogen storage and utilization in cells.
Modulation of Cell Cycle Progression: Akt signaling is involved in the regulation of cell cycle progression, promoting cell proliferation and preventing cell cycle arrest. Phosphatases in the Akt pathway modulate cell cycle progression by dephosphorylating Akt and inhibiting its activity. This dephosphorylation event can affect the phosphorylation status of downstream targets involved in cell cycle control, influencing the transition between cell cycle phases.
Tumor Suppressor Function: Phosphatases, particularly PP2A, act as tumor suppressors by regulating the Akt pathway. Dysregulation of phosphatase activity or expression is frequently observed in various cancers, leading to hyperactivation of Akt and uncontrolled cell growth and survival. Restoring phosphatase activity or enhancing their tumor suppressor function is an area of interest in cancer research, aiming to restore normal signaling and control aberrant Akt activation observed in cancer cells.
Regulation of Other Signaling Pathways: Phosphatases in the Akt pathway have broader implications beyond Akt regulation. They can modulate the activity of other signaling molecules and pathways involved in cell growth, proliferation, and apoptosis. For example, PP2A can dephosphorylate and regulate glycogen synthase kinase 3 (GSK3), mitogen-activated protein kinases (MAPKs), and other protein kinases involved in cell cycle control and cellular responses to stress.
Understanding the functions of phosphatases in the Akt pathway is essential for unraveling their roles in normal physiology, disease development, and potential therapeutic interventions.
Available Resources for Phosphatases in the Akt Pathway
Phosphatases play a key regulatory role in the Akt pathway. They regulate the activity of cell signaling pathways by removing phosphorylation modifications of Akt proteins. As important regulators in the Akt pathway, phosphatases play an important role in the regulation of cell proliferation, survival and metabolism.
Creative BioMart offers a wide range of products related to phosphatases in the Akt pathway, including recombinant proteins, cell and tissue lysates, and protein pre-coupled magnetic beads for research and experimental applications.
In addition to our existing products, we also provide customized services for specific research needs, including custom protein expression and purification. Our professional team will provide you with customized solutions based on your needs and experimental design.
Below are the molecules/targets related to Phosphatase in Akt pathway, click on them for product details. If you have any questions or requests, please feel free to contact us.
Why Choose Us?
Creative BioMart is committed to providing researchers with high quality products and services with extensive experience and specialized technical support. Our products meet stringent quality control standards and can support a wide range of research applications. Whether you are conducting basic science research or engaged in drug discovery, we can provide you with strong support and assistance.
We understand the importance of phosphatases in the Akt pathway, so our goal is to provide researchers with the highest quality, most reliable products and services to help them carry out their research in this area.
Reference:
- Zhang Q, Claret FX. Phosphatases: the new brakes for cancer development?. Enzyme Res. 2012;2012:659649. doi:10.1155/2012/659649

