CBR1
-
Official Full Name
carbonyl reductase 1 -
Overview
Carbonyl reductase is one of several monomeric, NADPH-dependent oxidoreductases having wide specificity for carbonyl compounds. This enzyme is widely distributed in human tissues. Another carbonyl reductase gene, CRB3, lies close to this gene on chromosome 21q. -
Synonyms
CBR1;carbonyl reductase 1;CBR;carbonyl reductase [NADPH] 1;SDR21C1;short chain dehydrogenase/reductase family 21C;member 1;carbonyl reductase (NADPH) 1;prostaglandin 9-ketoreductase;prostaglandin-E(2) 9-reductase;NADPH-dependent carbonyl reducta
Recombinant Proteins
- Human
- Rat
- Chicken
- Zebrafish
- Mouse
- Magnaporthe oryzae
- Cryptococcus Neoformans Var. Neoformans Serotype D
- Aspergillus Niger
- Aspergillus terreus
- Aspergillus oryzae
- S.cerevisiae
- Emericella nidulans
- Scheffersomyces stipitis
- Neurospora Crassa
- Schizosaccharomyces pombe
- Yarrowia lipolytica
- Mortierella alpina (Oleaginous fungus) (Mortierella renispora)
- Lodderomyces elongisporus
- Aspergillus clavatus
- Neosartorya fumigata
- Coccidioides immitis
- Vanderwaltozyma polyspora
- Ustilago maydis
- Candida albicans
- Ajellomyces capsulatus
- Neosartorya fischeri
- Meyerozyma guilliermondii
- E.coli
- Mammalian Cells
- HEK293
- Yeast
- In Vitro Cell Free System
- Non
- His
- DDK
- Myc
- GST
- Flag
- Avi
- Fc
Background
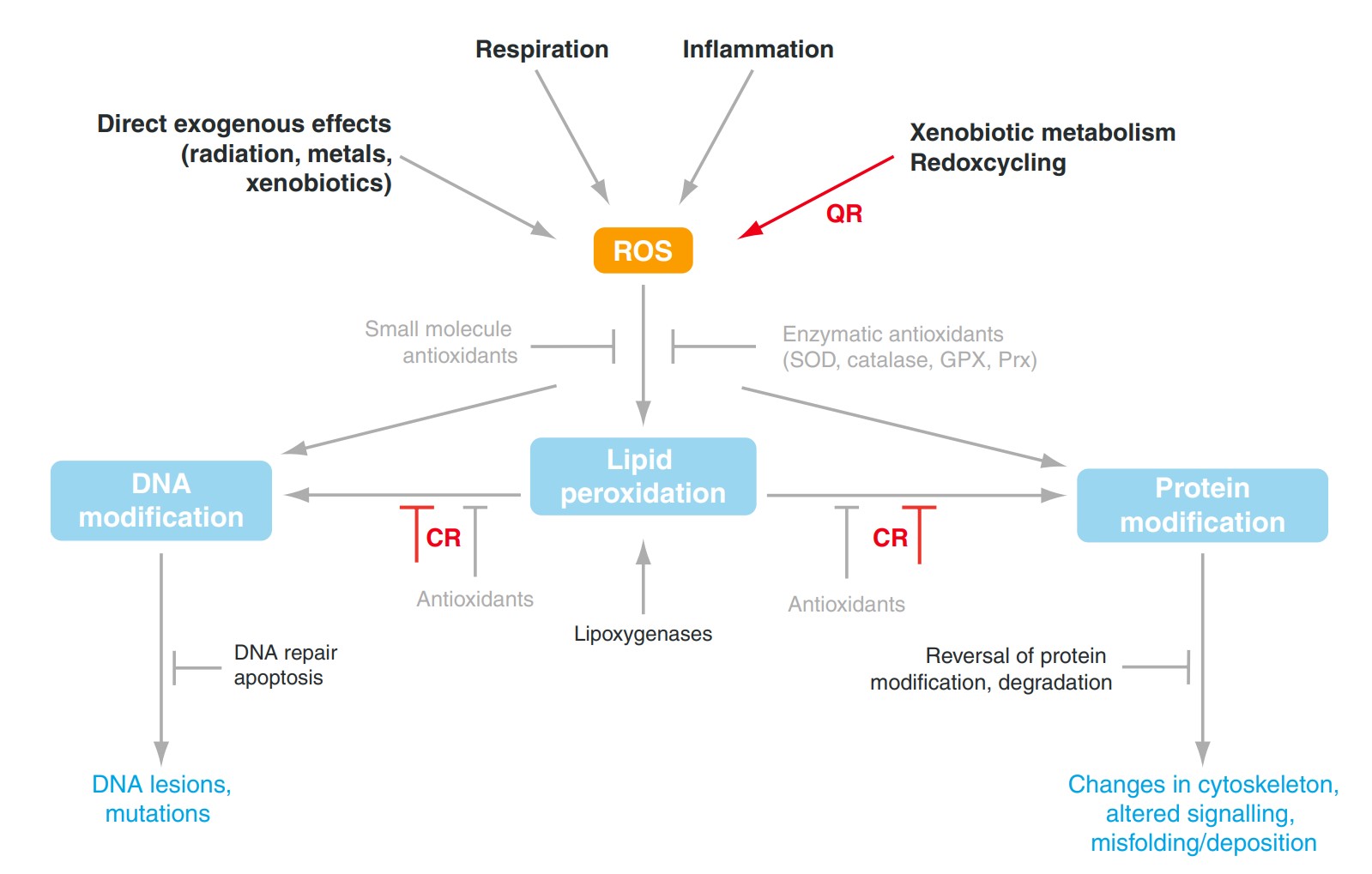
Fig1. Schematic overview on the involvement of quinone (QR) and carbonyl reductases (CR) in oxidative stress. (Udo Oppermann, 2007)
What is CBR1 protein?
CBR1 (carbonyl reductase 1) gene is a protein coding gene which situated on the long arm of chromosome 21 at locus 21q22. CBR1(Carbonyl reductase 1) is one of several monomericNADPH-dependentoxidoreductaseshaving wide specificity for carbonyl compounds. CBR1 is widely distributed in human tissues. CBR1 metabolizes many toxic environmentalquinonesand pharmacological relevant substrates such as the anticancerdoxorubicin. CBR1 can also convert prostaglandin E2 to prostaglandin F2-alpha. The CBR1 protein is consisted of 277 amino acids and its molecular mass is approximately 30.4 kDa.
What is the function of CBR1 protein?
CBR1 protein is a NADPH-dependent REDOX enzyme involved in the reduction of carbon-based compounds in living organisms. It can catalyze the reduction of aldehydes, ketones and other carbonyl compounds to produce corresponding alcohols. This process has several physiological functions in the organism, including detoxification, drug metabolism, fatty acid synthesis, and glucose metabolism. The expression and activity of CBR1 protein in cells are regulated by many factors, such as oxidative stress, hormone levels and nutritional status.
CBR1 Related Signaling Pathway
CBR1 protein is involved in several important signaling pathways, including nitric oxide (NO) signaling pathway, nuclear factor κB (NF-κB) signaling pathway, NADPH oxidase (NOX) signaling pathway, etc. In these pathways, CBR1 protein regulates intracellular REDOX balance and cell signal transduction through its reductase activity or binding to downstream signaling molecules, thereby affecting cell growth, apoptosis, differentiation and other life processes.
CBR1 Related Diseases
In genetic diseases, mutations in the CBR1 gene can lead to reduced or missing enzyme activity, which in turn affects intracellular metabolic pathways and causes disease. For example, CBR1 defects are associated with certain inherited metabolic diseases, such as mitochondrial lipid deposition disorders and fatty acid oxidation disorders. In environmental and lifestyle-related diseases, CBR1 proteins may influence the onset and progression of disease by regulating intracellular REDOX balance and detoxification responses. For example, changes in CBR1 expression and activity have been linked to cancer, diabetes, obesity, and cardiovascular disease. In addition, CBR1 is also involved in drug metabolism and toxicity, which may affect drug efficacy and safety.
Bioapplications of CBR1
Because CBR1 has a wide range of substrate specificity and can catalyze the reduction of a variety of aldehydes and ketones, it is used in the field of industrial biotechnology to produce chiral alcohols, which are of great value in pharmaceutical and fine chemicals. In the study of drug metabolism, the role of CBR1 helps to understand the process of drug disposal in the body, thus guiding drug design and optimization.
Case Study
Case study 1: Quinn A Best, 2016
Quinoneoxidoreductase isozyme-1 within human cells, based on results from an investigation of the stability of various rhodamines and seminaphthorhodamines toward the biological reductant NADH, present at ~100-200 μM within cells. While rhodamines are generally known for their chemical stability, the researchers observe that NADH causes significant and sometimes rapid modification of numerous rhodamine analogues, including those oftentimes used in imaging applications. A relationship between the structural features of the rhodamines and their reactivity with NADH is observed. Rhodamines with increased alkylation on the N3- and N6-nitrogens, as well as the xanthene core, react the least with NADH; whereas, nonalkylated variants or analogues with electron-withdrawing substituents have the fastest rates of reaction. These outcomes allowed us to judiciously construct a seminaphthorhodamine-based, turn-on fluorescent probe that is capable of selectively detecting the cancer-associated, NADH-dependent enzyme.
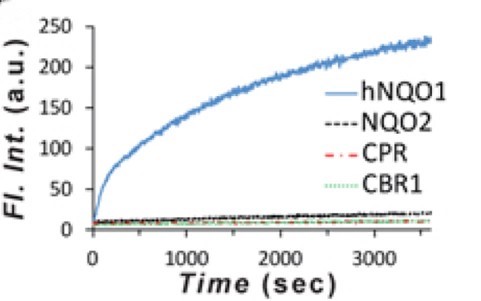
Fig1. Activation of 5 μM probe by hNQO1, in contrast to a lack thereof by other reductase enzymes (human).
Case study 2: Miyong Yun, 2018
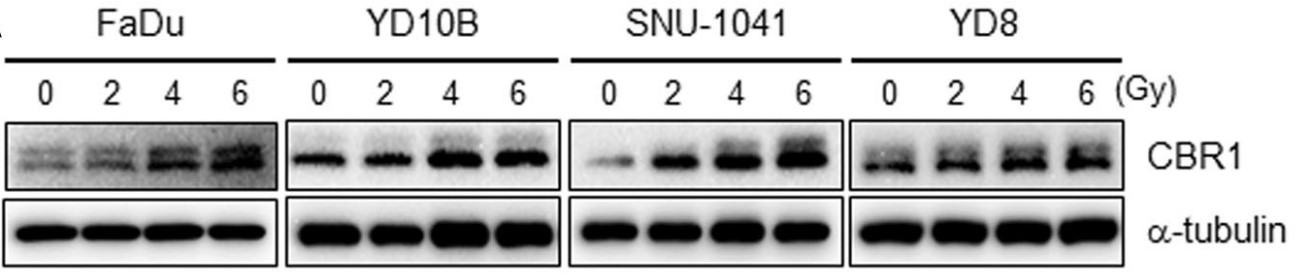
Human carbonyl reductase 1 (CBR1) plays major roles in protecting cells against cellular damage resulting from oxidative stress. Although CBR1-mediated detoxification of oxidative materials increased by stressful conditions including hypoxia, neuronal degenerative disorders, and other circumstances generating reactive oxide is well documented, the role of CBR1 under ionising radiation (IR) is still unclear.
The formalin-fixed and paraffin-embedded tissues of 85 patients with head and neck squamous cell carcinoma (HNSCC) were used to determine if CBR1 expression effects on survival of patients with treatment of radiotherapy. Subsequently colony formation assays and xenograft tumor mouse model was used to verify the relationship between CBR1 expression and radiosensitivity in HNSCC cells. To verify CBR1-mediated molecular signalling pathways, cell survival, DNA damage/repair, reactive oxygen species (ROS), cell cycle distribution and mitotic catastrophe in HNSCC cells with modulated CBR1 expression by knockdown or overexpression were measured using by colony formation assays, flow cytometry, qRT-PCR and western blot analysis. CBR1 has a key role in DNA damage response through regulation of IR-mediated ROS generation. Consistently, CBR1 expression is highly correlated with patient survival after and susceptibility to radiation therapy.
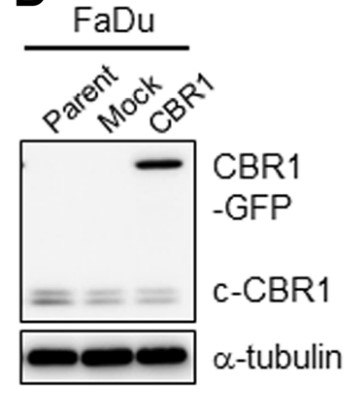
Fig3. CBR1 protein expression was evaluated in FaDu and YD38 cells transfected with Mock and CBR1/WT vectors.
Quality Guarantee
High Purity

Fig1. SDS-PAGE (CBR1-0467H) (PROTOCOL for western blot)
.
Fig2. SDS-PAGE (CBR1-591H) (PROTOCOL for western blot)
Involved Pathway
CBR1 involved in several pathways and played different roles in them. We selected most pathways CBR1 participated on our site, such as Arachidonic acid metabolism,Metabolism of xenobiotics by cytochrome P,Metabolic pathways, which may be useful for your reference. Also, other proteins which involved in the same pathway with CBR1 were listed below. Creative BioMart supplied nearly all the proteins listed, you can search them on our site.
| Pathway Name | Pathway Related Protein |
|---|---|
| Metabolic pathways | AKR1B1,HK3,CERS5,TKTB,ACSM3,MVDA,PAFAH1B1A,GGT7,NDUFB2,MAT1A |
| Arachidonic acid metabolism | PTGS2,CYP2J5,CYP2AD3,CYP2P6,PTGESL,CBR3,PLA2G5,AKR1C21,PLA2G4B,CYP1B1 |
| Chemical carcinogenesis | CYP2E1,CYP2C55,ALDH3A1,CYP2C70,GSTA5,GSTM3,CYP3A41B,SULT1A2,GSTP2,ADH2-2 |
| Metabolism of xenobiotics by cytochrome P | UGT2A2,CBR2,GSTP2,HSD11B1,GSTM3,UGT1A9,GSTO1,GSTM,GSTP1,GSTA1 |
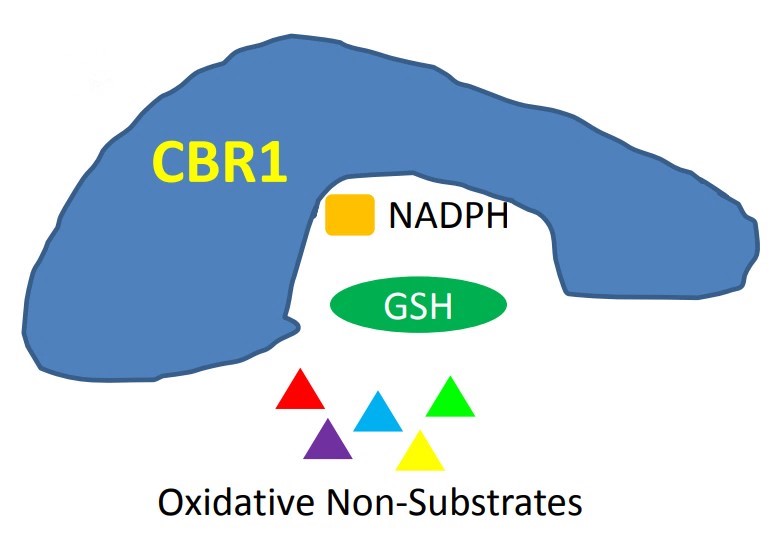
Fig1. When there is no substrate binding to CBR1, GSH protects the active site and NADPH from attacks of oxidative small molecules. (Sophia M Shi, 2017)
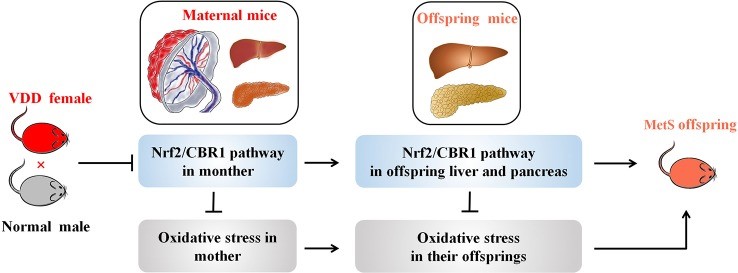
Fig2. A working model illustrating the effect of maternal VDD on MetS in their offspring. (Jianqiong Zheng, 2020)
Protein Function
CBR1 has several biochemical functions, for example, 15-hydroxyprostaglandin dehydrogenase (NADP+) activity,carbonyl reductase (NADPH) activity,oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor. Some of the functions are cooperated with other proteins, some of the functions could acted by CBR1 itself. We selected most functions CBR1 had, and list some proteins which have the same functions with CBR1. You can find most of the proteins on our site.
| Function | Related Protein |
|---|---|
| oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | AKR1C6,NDUFS7,DCXR,AKR1C4,AKR1C21,AKR1C2,AKR1C3,DHRS4 |
| carbonyl reductase (NADPH) activity | DHRS2,CBR3,CBR2,DHRS4 |
Interacting Protein
CBR1 has direct interactions with proteins and molecules. Those interactions were detected by several methods such as yeast two hybrid, co-IP, pull-down and so on. We selected proteins and molecules interacted with CBR1 here. Most of them are supplied by our site. Hope this information will be useful for your research of CBR1.
PRKAB1;EIF1B;EPB41;VDAC1;NME2;MCC;HLA-B;HLA-C;EIF6;GABARAPL2;IKBKE;ATG101;DDA1;UBA5;FTSJ1;TNIK;TRAF6;EGFR;ERCC8
Resources
Related Services
Related Products
References




