Synaptic Adhesion Molecules
Related Symbol Search List
- CLSTN1
- CLSTN2
- CLSTN3
- CNTN1
- CNTN3
- CNTN4
- CNTN5
- CADM1
- CADM2
- CADM4
- NLGN1
- NLGN2
- NLGN3
- NLGN4X
- NRXN2
- NRXN3
- CBLN1
- CBLN2
- CBLN4
- CDH12
- DSCAM
- EFNA4
- EFNA5
- EFNB1
- Efnb2
- EPHA5
- L1CAM
- LRFN1
- LRFN3
- LRFN4
- LSAMP
- NCAM1
- NEGR1
- NPTN
- OPCML
- PCDH8
- Syndecan-2
- SLITRK1
- SLITRK2
- SLITRK4
- SLITRK6
Immunology Background
About Synaptic Adhesion Molecules
Synaptic adhesion molecules are a diverse group of proteins that play crucial roles in the formation and maintenance of connections, or synapses, between neurons in the brain. These molecules are located on the surface of both pre- and postsynaptic neurons, where they serve as key molecular bridges between these two compartments.
There are several families of synaptic adhesion molecules, including neurexins, neuroligins, cadherins, immunoglobulin superfamily (IgSF) proteins, and leucine-rich repeat transmembrane (LRRTM) proteins, among others. These molecules are characterized by their structure, which typically consists of extracellular domains that mediate protein-protein interactions, as well as cytoplasmic domains that can interact with the intracellular signaling machinery.
The main function of synaptic adhesion molecules is to promote synapse formation by mediating cell-cell adhesion between presynaptic and postsynaptic neurons. They do this by binding to specific partners across the synapse, such as receptors, scaffolding proteins, and cytoskeletal components. In this way, they provide structural stability to synapses and contribute to the localization and organization of synaptic components.
In addition to their role in synapse formation, synaptic adhesion molecules are also involved in synapse specification and maturation, synaptic plasticity, and synapse elimination. They regulate the development and refinement of neural circuits by controlling synapse number, strength, and specificity.
The dysfunction or altered expression of synaptic adhesion molecules has been implicated in various neurological disorders, including autism spectrum disorders, schizophrenia, and intellectual disabilities. Studies have shown that mutations in genes encoding these molecules can disrupt synaptic connectivity and impair brain function, highlighting their importance in normal brain development and function.
Biological Functions of Synaptic Adhesion Molecules
Synaptic adhesion molecules are involved in various biological functions that contribute to synapse formation, synaptic plasticity, and proper neuronal circuitry. Here are some key biological functions of synaptic adhesion molecules:
Synapse Formation and Stabilization: Synaptic adhesion molecules are essential for the initial formation of synapses during development. They mediate the recognition and adhesion between pre- and postsynaptic neurons, promoting the assembly of synaptic components. These molecules facilitate the precise alignment of pre- and postsynaptic compartments, allowing for the establishment of functional synaptic connections. Additionally, synaptic adhesion molecules contribute to the stabilization of synapses by promoting the clustering of neurotransmitter receptors and other synaptic proteins at the synaptic junction.
Synaptic Differentiation: Synaptic adhesion molecules participate in the process of synaptic differentiation, which involves the maturation and specialization of synapses. They contribute to the organization and segregation of pre- and postsynaptic components, ensuring the proper localization of neurotransmitter release machinery, postsynaptic receptors, and scaffolding proteins. This organization is crucial for the efficient transmission of signals across synapses and the establishment of synaptic specificity.
Synaptic Plasticity: Synaptic adhesion molecules play a role in synaptic plasticity, the ability of synapses to undergo activity-dependent changes in strength and structure. These molecules can modulate synaptic strength by regulating the number and function of neurotransmitter receptors at the postsynaptic membrane. They can also influence the formation and remodeling of synaptic connections in response to synaptic activity, contributing to the structural and functional plasticity of neural circuits.
Neuronal Network Development: Synaptic adhesion molecules are involved in the development of neuronal networks. They participate in processes such as axon guidance, dendritic branching, and target selection, which are crucial for establishing proper connectivity between neurons. By mediating cell-cell interactions and adhesive interactions between neurons, these molecules contribute to the intricate wiring of the nervous system during development.
Synaptic Homeostasis: Synaptic adhesion molecules play a role in maintaining synaptic homeostasis, the ability of synapses to maintain stable and balanced neurotransmission. They contribute to the regulation of synaptic strength, ensuring that synaptic activity remains within an optimal range. Synaptic adhesion molecules can modulate the availability and localization of neurotransmitter receptors, facilitating the fine-tuning of synaptic transmission.
Synaptic Modularity: Synaptic adhesion molecules contribute to the organization of synapses into functional modules or subtypes. Different adhesion molecules are expressed in specific synaptic subtypes, allowing for the formation of specialized synapses with distinct properties. This modularity enables the diversification and specialization of synaptic circuits, contributing to the functional diversity observed in the nervous system.
Neurodevelopmental Disorders: Defects or alterations in synaptic adhesion molecules have been associated with various neurodevelopmental disorders, including autism spectrum disorders (ASDs) and intellectual disabilities. Dysregulation of these molecules can disrupt synaptic connectivity, impair synaptic function, and lead to abnormal neuronal circuitry. Understanding the roles of synaptic adhesion molecules in neurodevelopment is crucial for elucidating the mechanisms underlying these disorders and developing targeted therapeutic strategies.
Overall, synaptic adhesion molecules play fundamental roles in synapse formation, synaptic plasticity, synaptic organization, and neuronal circuit development. They contribute to the establishment and maintenance of functional synapses, ensuring proper neuronal communication and the integrity of neural circuits.
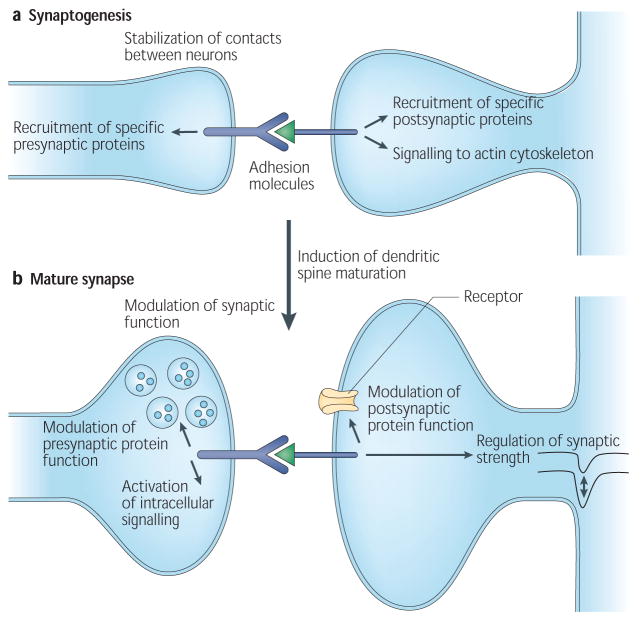 Fig.1 Cell-adhesion molecules function in synapse. (Dalva MB, 2007)
Fig.1 Cell-adhesion molecules function in synapse. (Dalva MB, 2007)
Cell-adhesion molecules (CAMs) function throughout the life of a synapse. They are important during synaptogenesis, but also in the mature synapse.
The Application Areas of Synaptic Adhesion Molecules
Neurodevelopmental Disorders: Synaptic adhesion molecules are implicated in several neurodevelopmental disorders, including autism spectrum disorders (ASDs) and intellectual disabilities. Research focuses on studying the role of specific synaptic adhesion molecules in these disorders to understand their contribution to altered synaptic connectivity and function. This knowledge can lead to the development of diagnostic biomarkers and potential therapeutic interventions targeting synaptic adhesion molecules.
Synaptic Plasticity and Learning: Synaptic adhesion molecules are involved in synaptic plasticity, which underlies learning and memory processes. Researchers investigate the function of these molecules in mediating changes in synaptic strength and structural modifications during learning and memory formation. Understanding the role of synaptic adhesion molecules in synaptic plasticity can provide insights into the mechanisms underlying cognitive processes and inform strategies for enhancing learning and memory.
Neurodegenerative Disorders: Synaptic adhesion molecules are implicated in neurodegenerative disorders such as Alzheimer's disease and Parkinson's disease. Research focuses on studying alterations in synaptic adhesion molecules associated with these disorders and their impact on synaptic dysfunction and neuronal degeneration. Targeting synaptic adhesion molecules may offer potential therapeutic avenues for neurodegenerative diseases by restoring synaptic connectivity and function.
Neural Regeneration and Repair: Synaptic adhesion molecules play a crucial role in synaptic regeneration and repair following neural injury or damage. Researchers investigate the involvement of these molecules in axonal growth, synapse formation, and functional recovery after injury. Understanding the mechanisms by which synaptic adhesion molecules promote neural regeneration can guide the development of therapies to enhance neural repair in conditions such as spinal cord injury and stroke.
Drug Discovery and Therapeutics: Synaptic adhesion molecules represent potential targets for drug discovery and therapeutics. Modulating the function of these molecules may offer opportunities to modulate synaptic connectivity, plasticity, and function. Researchers explore the development of small molecules, peptides, or antibodies that can selectively interact with synaptic adhesion molecules to restore synaptic deficits in neurological disorders or enhance cognitive function.
Biomaterials and Tissue Engineering: Synaptic adhesion molecules are utilized in the field of biomaterials and tissue engineering to guide the regeneration and organization of neural circuits. Incorporating specific adhesion molecules into biomaterial scaffolds can facilitate the formation of functional synapses and promote neural network development. This approach holds promise for applications in neural tissue regeneration, implantable devices, and neuroprosthetics.
Diagnostic and Prognostic Tools: Alterations in synaptic adhesion molecules can serve as diagnostic and prognostic markers for various neurological disorders. Researchers investigate changes in the expression and localization of these molecules in patient samples to identify potential biomarkers associated with disease progression or treatment response. Such markers can aid in early diagnosis, monitoring disease progression, and assessing treatment efficacy.
The application areas of synaptic adhesion molecules are diverse and span from basic research to clinical applications. They contribute to our understanding of neurodevelopmental disorders, synaptic plasticity, neurodegenerative diseases, neural regeneration, and therapeutic interventions. Continued research on synaptic adhesion molecules holds promise for advancing our knowledge of synaptic connectivity, developing targeted treatments for neurological disorders, and improving neural repair strategies.
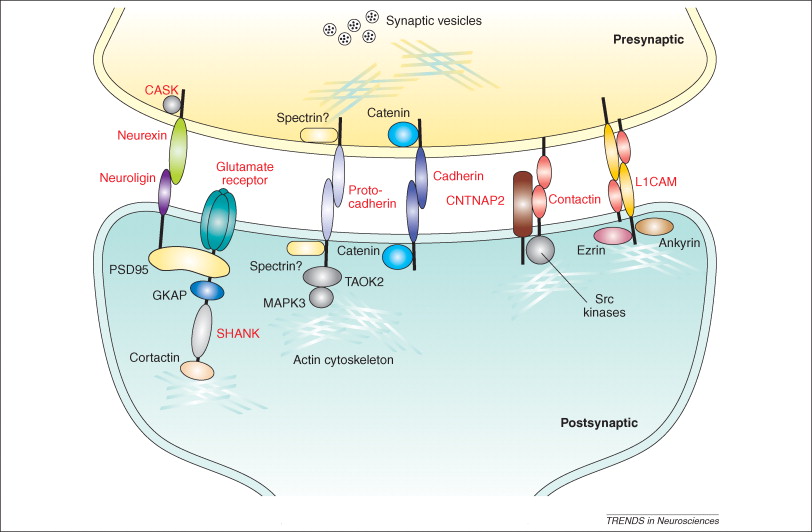 Fig.2 Synaptic cell-adhesion molecules. (Betancur C, 2009)
Fig.2 Synaptic cell-adhesion molecules. (Betancur C, 2009)
Different families of cell-adhesion molecules (CAMs) in the synapse. CAMs are divided in groups based on their structure,function and localization.
Available Resources for Synaptic Adhesion Molecules
Creative BioMart provides products related to synaptic adhesion molecules. The product line includes recombinant proteins, cell and tissue lysates, protein pre-coupled magnetic beads, and others, aiming to support your research in the field of synaptic adhesion molecules and related Scientific exploration. Our custom services are designed to meet your specific research needs, including protein expression, purification and custom development. In addition, we offer a wide range of educational and technical resources, including application guides and protocols, designed to help improve your understanding of synaptic adhesion molecules and improve the success of your research experiments.
Feel free to view the synaptic adhesion-related molecules below and click for more comprehensive resources. For more details or if you have additional questions, please feel free to contact us.
References:
- Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32(7):402-412. doi:10.1016/j.tins.2009.04.003.
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8(3):206-220. doi:10.1038/nrn2075

