Active Recombinant E. coli RNase H
| Cat.No. : | rnh-63E |
| Product Overview : | A recombinant?E. coli?strain carrying the RNAse H (rnh) gene from?E.coli. |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | E.coli |
| Source : | E.coli |
| Tag : | Non |
| Description : | E.coli?RNase H (rnh) is an endoribonuclease which degrades the RNA strand of RNA/DNA hybrid molecules. RNase H digestion produces ribonucleotide molecules with 5-phosphate and 3-hydroxyl termini. RNAse H is nearly inactive against single or double-stranded RNA molecules. |
| Bio-activity : | 625,000 U/mg |
| Purity : | >99% by SDS-PAGE |
| Unit Definition : | 1 unit is defined as the amount of enzyme that will hydrolyze 1 nmol of RNA from an 3H-labeled DNA:RNA hybrid molecule into acid-soluble material in 20 minutes at 37°C. |
| Storage : | -25 to -15°C |
| Concentration : | 5,000 U/ml |
| Storage Buffer : | Supplied in: 20 mM Tris-HCl, 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 50% glycerol, pH 7.9 @ 25°C Supplied with: 10X RNAse H Buffer: 500mM Tris-HCl, 750 mM KCl, 30 mM MgCl2, 100 mM DTT, pH 8.3 @ 25?C. |
| ◆ Recombinant Proteins | ||
| rnh-63E | Active Recombinant E. coli RNase H | +Inquiry |
Characterization of Escherichia coli RNase H Discrimination of DNA Phosphorothioate Stereoisomers
Journal: Nucleic Acid Therapeutics PubMed ID: 34619060 Data: 2021/12/1
Authors: ?ukasz J. Kie?piński, Erik Daa Funder, Peter H. Hagedorn
Article Snippet:Gel electrophoresis was performed in 1 × TBE buffer on 15% TBE-Urea gel (Novex ? ) with constant 180 V. Bands were visualized using ChemiDoc Touch Imaging System (Bio-Rad) on Blue Sample Tray.Gel electrophoresis was performed in 1 × TBE buffer on 15% TBE-Urea gel (Novex ? ) with constant 180 V. Bands were visualized using ChemiDoc Touch Imaging System (Bio-Rad) on Blue Sample Tray.. The recombinant E. coli RNase H enzyme used in the experiments was purchased from Creative BioMart (cat. RNASEH1-433H).. It was provided as a solution between 20 and 60 U/μL, but for the concentrations reported below, it was assumed to be 60 U/μL.It was provided as a solution between 20 and 60 U/μL, but for the concentrations reported below, it was assumed to be 60 U/μL.
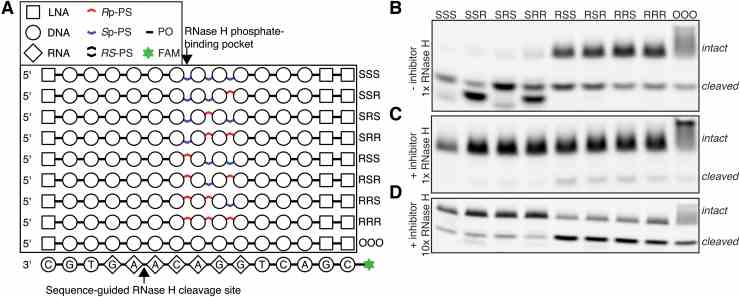
Characterization of
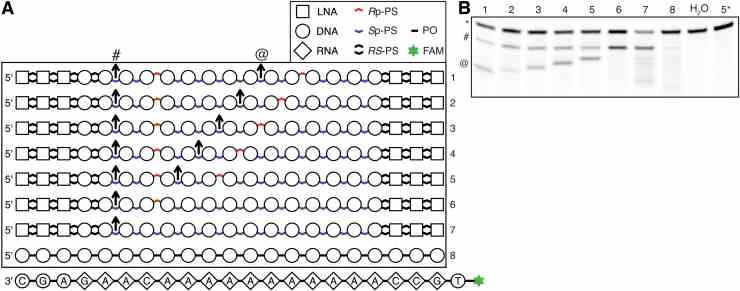
Control of
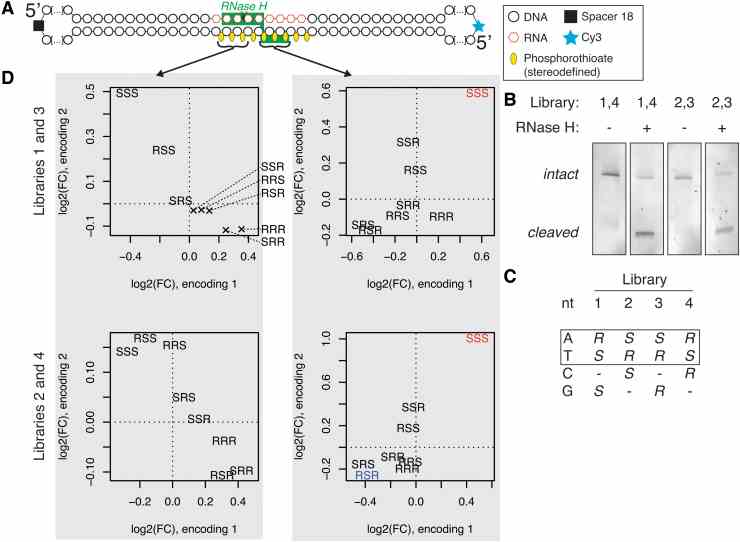
Massive parallel screen for preferred chiral motifs. (A) Structure of a circular substrate used in the experiment, with drawing of one of the possible
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All rnh Products
Required fields are marked with *
My Review for All rnh Products
Required fields are marked with *



