Ionic Detergents for Active Membrane Protein Isolation: Principles, Strategies, and Trade-offs
Ionic detergents represent a double-edged sword in membrane protein solubilization, offering unparalleled extraction efficiency at the risk of structural denaturation and functional inactivation. This article systematically examines the molecular mechanisms, strategic selection frameworks, and activity-preserving methodologies essential for successful membrane protein isolation. We dissect critical parameters including Critical Micellar Concentration (CMC), micelle dimensions, and charge-mediated interference, with particular emphasis on their profound impacts on functional proteomics studies. Through detailed comparative analyses, case studies, and troubleshooting protocols, we provide researchers with actionable guidelines to navigate the inherent yield-functionality trade-off. The review culminates in forward-looking perspectives on rational detergent design, AI-driven screening, and emerging detergent-free alternatives.
Introduction: The Dilemma of Ionic Detergents in Membrane Protein Research
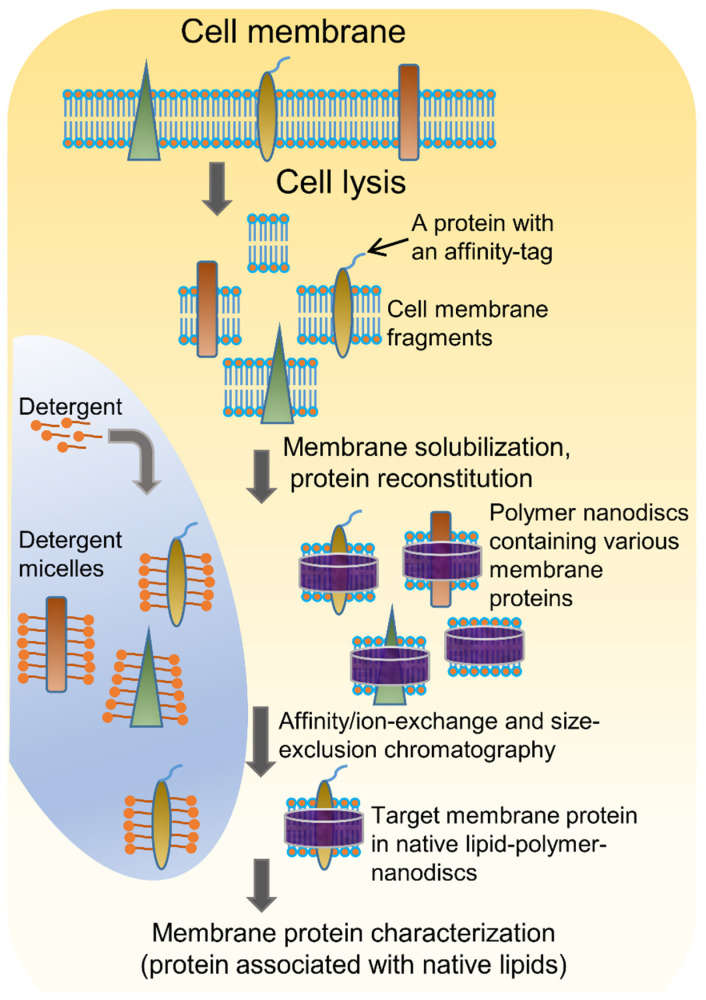 Fig1. Schematic representation of membrane protein purification using the traditional detergent-based approach and the detergent-free polymer-based approach.
Fig1. Schematic representation of membrane protein purification using the traditional detergent-based approach and the detergent-free polymer-based approach.The Core Challenge of Membrane Protein Solubilization
Membrane proteins constitute approximately 30% of the proteome and mediate crucial cellular functions including signal transduction, molecular transport, and energy conversion. Their hydrophobic transmembrane domains, typically spanning 20-25 Å through the lipid bilayer, necessitate disruption of native membrane architecture for aqueous solubilization. This process demands meticulous preservation of tertiary and quaternary structures to maintain biological activity—a challenge that becomes exponentially complex when employing ionic detergents.
The fundamental paradox lies in the competing requirements: efficient lipid bilayer disruption versus minimal protein structural perturbation. While non-ionic detergents like dodecyl-β-D-maltoside (DDM) have become gold standards for functional studies, they often fail to extract proteins from dense membrane microdomains or insoluble aggregates. Ionic detergents, with their charged head groups, demonstrate superior solubilization kinetics but introduce significant risks of irreversible denaturation through charge-charge repulsion and disruption of native protein-lipid interactions.
Defining Ionic Detergents: Classification and Molecular Architecture
Ionic detergents are amphiphilic molecules comprising a hydrophobic alkyl chain (typically C8-C16) and a charged hydrophilic head group. They are categorized based on head group charge:
Anionic detergents:
- Sodium dodecyl sulfate (SDS): C₁₂H₂₅SO₄⁻Na⁺, the most potent denaturant
- Bile acid salts: Cholate, deoxycholate (DOC), and taurodeoxycholate with steroid ring structures
- Sarcosyl (N-lauroylsarcosine): Milder carboxylate head group
Cationic detergents:
- Cetyltrimethylammonium bromide (CTAB): C₁₆H₃₃N⁺(CH₃)₃Br⁻
- Dodecyltrimethylammonium bromide (DTAB)
Their CMC values range dramatically: SDS (8.2 mM), DOC (4-6 mM), CTAB (0.9 mM), directly impacting experimental optimization.
The Activity-Preservation Paradox
Ionic detergents are frequently classified as "harsh" or "denaturing" due to their propensity to disrupt not only lipid-lipid interactions but also protein-protein and protein-lipid contacts essential for native conformation. The charged head group competes with polar residues in transmembrane segments, potentially unfolding α-helices and β-barrels. However, their unmatched ability to solubilize recalcitrant proteins—including G-protein coupled receptors (GPCRs), transporters, and channel complexes—necessitates strategic deployment. The central tenet is not avoidance, but controlled application through concentration modulation, temperature control, and rapid downstream processing.
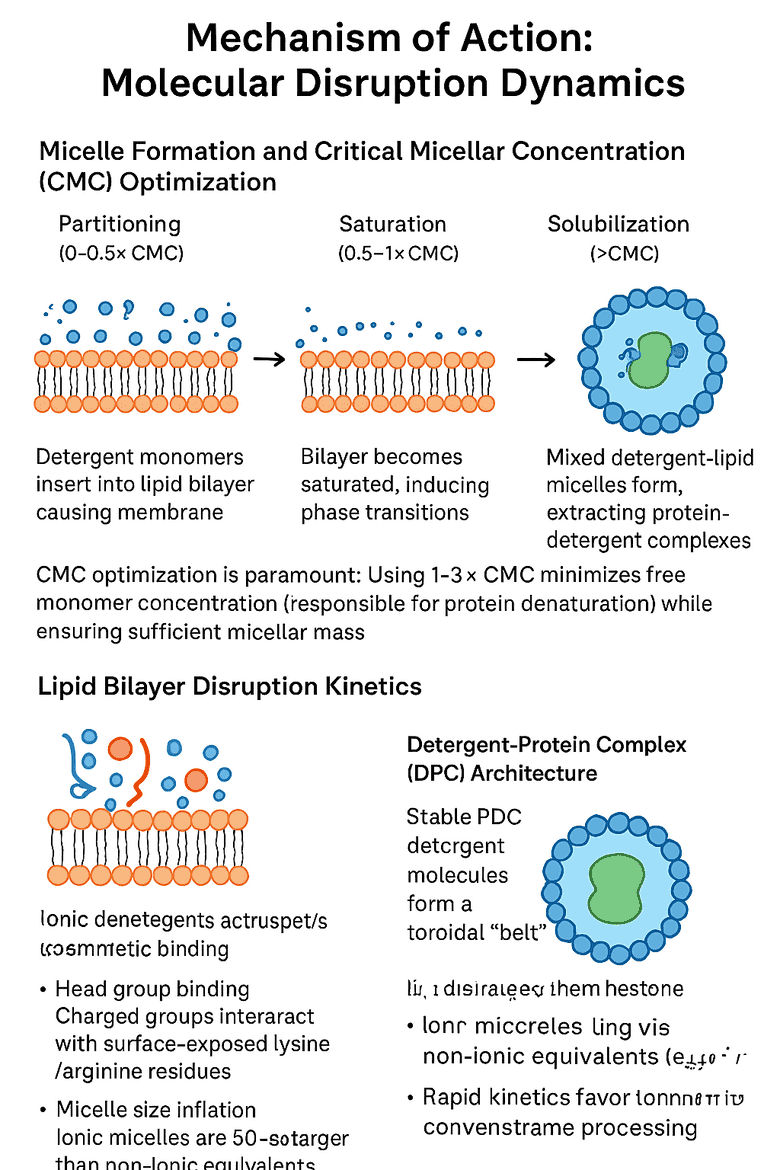
Mechanism of Action: Molecular Disruption Dynamics
Micelle Formation and Critical Micellar Concentration (CMC) Optimization
The solubilization process occurs in three distinct phases:
- Partitioning (0-0.5×CMC): Detergent monomers insert into the lipid bilayer, causing membrane thinning
- Saturation (0.5-1×CMC): Bilayer becomes saturated, inducing phase transitions
- Solubilization (>CMC): Mixed detergent-lipid micelles form, extracting protein-detergent complexes (PDCs)
CMC optimization is paramount: Using 1-3×CMC minimizes free monomer concentration (responsible for protein denaturation) while ensuring sufficient micellar mass for extraction. For SDS, this translates to 8.2-24.6 mM (0.24-0.72% w/v), whereas deoxycholate requires only 4-12 mM. The "minimal effective concentration" (MEC) principle dictates starting at 1×CMC and incrementing in 0.5×CMC steps until >80% extraction is achieved.
Lipid Bilayer Disruption Kinetics
Ionic detergents exhibit charge-mediated disruption fundamentally different from steric mechanisms of non-ionic agents. The charged head group electrostatically repels negatively charged phospholipid head groups (e.g., phosphatidylserine, phosphatidic acid), creating localized membrane destabilization. This is followed by hydrophobic tail insertion, which disorders acyl chain packing and reduces bilayer thickness by 10-15 Å.
Time-course studies reveal that SDS achieves complete solubilization within 5-10 minutes at 37°C, whereas milder deoxycholate may require 30-60 minutes at 4°C to prevent over-extraction. Rapid kinetics favor high-throughput processing but demand stringent temperature control to prevent thermal denaturation synergism.
Detergent-Protein Complex (DPC) Architecture
In a stable PDC, detergent molecules form a toroidal "belt" around the hydrophobic transmembrane region, typically at a detergent : protein molar ratio of 50:1 to 200:1. However, ionic detergents demonstrate asymmetric binding patterns:
- Head group binding: Charged groups interact with surface-exposed lysine/arginine residues
- Micelle size inflation: Ionic micelles are 30-50% larger than non-ionic equivalents (e.g., SDS micelle MW ~18 kDa vs. DDM ~50 kDa)
This size inflation critically impacts downstream size exclusion chromatography (SEC), causing proteins to elute with apparent molecular weights inflated by 60-100 kDa.
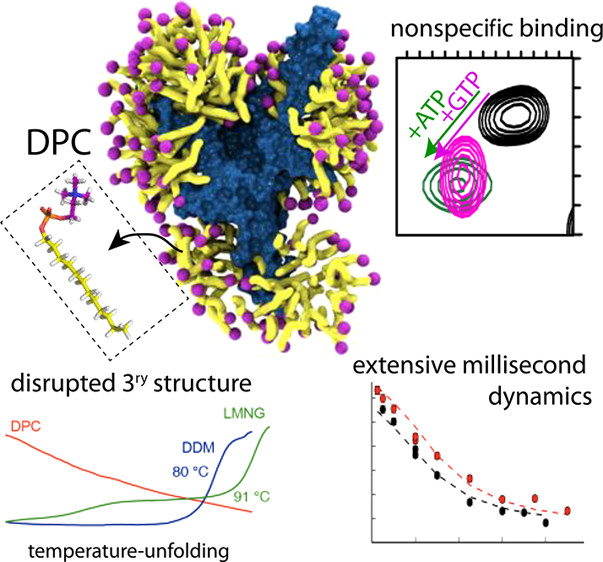 Fig2. Dodecylphosphocholine (DPC) is the most widely used detergent for MP structure determination by NMR (J. Phys. Chem. Lett. 2018,)
Fig2. Dodecylphosphocholine (DPC) is the most widely used detergent for MP structure determination by NMR (J. Phys. Chem. Lett. 2018,)Charge Effects on Protein Stability
The ionic nature introduces pH-dependent behavior: Below pH 7.4, carboxylate detergents (cholate, deoxycholate) become partially protonated, reducing effective charge and solubilization power. Conversely, CTAB's quaternary ammonium group remains fully charged across pH 2-12, providing consistent but potentially harsher extraction.
Charge shielding effects are also significant: At physiological ionic strength (150 mM NaCl), electrostatic repulsion between anionic detergents and negatively charged protein surfaces is partially screened, reducing unintended binding. However, high salt (>500 mM) can precipitate ionic detergents via counterion effects.
Commonly Used Ionic Detergents: Properties and Strategic Applications
Anionic Detergents in Detail
SDS (Sodium Dodecyl Sulfate)
- CMC: 8.2 mM (0.24% w/v, 25°C)
- Aggregation number: ~62 monomers per micelle
- Micelle molecular weight: 18 kDa
- Key applications:
- Proteomics: Irreversible denaturation for mass spectrometry and gel electrophoresis
- Inclusion body solubilization: Effective at 1-2% w/v for bacterial overexpression systems
- Membrane stripping: Complete lipid removal for analytical purposes
Critical limitation: SDS binds to proteins at ~1.4 g/g ratio, coating them with a uniform negative charge that prevents refolding. While excellent for analytical workflows, SDS is contraindicated for functional studies unless followed by exhaustive detergent exchange.
Deoxycholate (SDC) and Cholate
- CMC (DOC): 4-6 mM (0.21% w/v)
- Micelle size: Smaller toroidal structures (aggregation number ~4-10)
- Unique features: Steroid ring structure provides facial amphiphilicity—one side hydrophobic, the other hydrophilic
Advantages for functional work:
- Relative mildness: Can preserve GPCR signaling activity when used at sub-CMC concentrations (2-4 mM)
- High CMC: Facilitates removal by dialysis or dilution
- Versatility: Compatible with certain ion-exchange chromatography at low concentrations (<2 mM)
Protocol example: For rhodopsin extraction, 20 mM HEPES pH 7.5 buffer containing 1% DOC (w/v), 100 mM NaCl, and complete protease inhibitors, incubated at 4°C for 30 minutes with gentle agitation, yielded active receptor capable of light-activated conformational changes.
Sarcosyl (N-Lauroylsarcosine)
- CMC: 14 mM
- Milder profile: Carboxylate head group with reduced denaturing tendency
- Specialized use: Selective solubilization of outer membrane proteins from Gram-negative bacteria while leaving inner membrane proteins intact
Cationic Detergents: CTAB and Alternatives
CTAB (Cetyltrimethylammonium Bromide)
- CMC: 0.9 mM (0.033% w/v)
- Micelle molecular weight: 62 kDa
- Strategic applications:
- Acidic phospholipid-rich membranes: Effective for mitochondrial inner membrane (cardiolipin content >20%)
- DNA-protein complex disruption: For transcription factor studies
Severe limitation: CTAB binds irreversibly to silica-based chromatography resins and forms insoluble complexes with nucleic acids. Requires extensive buffer exchange before any chromatographic step. Additionally, its low CMC makes removal extremely difficult, often necessitating detergent adsorption onto hydrophobic beads (Bio-Beads SM-2).
Comparative Parameter Table
| Detergent | Type | CMC (mM) | Micelle MW (kDa) | Denaturing Index | Compatible Methods | Removal Difficulty |
|---|---|---|---|---|---|---|
| SDS | Anionic | 8.2 | 18 | Very High | SDS-PAGE, MS | Very Low (dialysis) |
| DOC | Anionic | 5.0 | 2-5 | Medium | SEC (limited), IMAC | Low (dialysis) |
| CTAB | Cationic | 0.9 | 62 | High | Precipitation only | Very High (adsorption) |
| FC-14 | Zwitterionic | 0.2 | 50 | Low | All chromatography | Medium |
Strategic Considerations for Active Protein Isolation
The Golden Rules of Detergent Screening
A systematic screening workflow is essential:
Step 1: HLB Value Optimization
The Hydrophilic-Lipophilic Balance (HLB) predicts extraction efficiency. For membrane proteins, optimal HLB ranges from 12-15. Deoxycholate (HLB ~16) shows superior performance over cholate (HLB ~18) for GPCRs, extracting 40-60% more functional receptor.
Step 2: Concentration Titration
Perform small-scale (1 mL) extractions at detergent concentrations of:
- 0.5×CMC (baseline)
- 1×CMC (recommended start)
- 2×CMC (maximum for functional work)
- 5×CMC (for analytical/structural studies)
Assess both solubilization efficiency (Western blot quantification) and activity retention (ligand binding, enzymatic assay). Plotting these parameters reveals the sweet spot where extraction plateaus but activity remains >70% of native membrane levels.
Step 3: Buffer Composition Fine-Tuning
- Salt concentration: 100-200 mM NaCl optimizes charge shielding without precipitating detergent
- pH: Match detergent pKa (7-8 for carboxylates) to maintain full ionization
- Reducing agents: 1-5 mM DTT prevents aggregation but can reduce SDS effectiveness
Sequential Solubilization: A Multi-Tiered Approach
This advanced strategy maximizes both yield and functionality:
Phase 1 (Gentle extraction): 1% DDM or 1% digitonin for 1 hour at 4°C → isolate "easily extractable" active pool Phase 2 (Moderate ionic): Supernatant from Phase 1 re-extracted with 2-4 mM deoxycholate → capture moderately recalcitrant proteins Phase 3 (Harsh ionic): Pellet from Phase 2 solubilized with 1% SDS → recover insoluble aggregates for structural analysis
This tiered approach segregates protein populations based on membrane embedding depth and lipid microenvironment, preserving the most delicate complexes for functional assays.
Temperature and Temporal Control
Temperature profoundly influences outcomes:
- 4°C: Reduces denaturation kinetics by 5-10 fold, but increases required incubation time to 30-60 minutes
- Room temperature (22°C): Balanced approach for most ionic detergents, 15-30 minutes
- 37°C: Accelerates solubilization but dramatically increases denaturation risk; only recommended for SDS-based analytical extractions
Time-course experiment: Deoxycholate extraction of the serotonin transporter (SERT) showed maximal activity at 20 minutes (4°C), with 50% activity loss by 60 minutes due to progressive delipidation.
Protease Inhibitor Cocktails
Ionic detergents can activate latent proteases by disrupting compartmentalization. Implement broad-spectrum protection:
- 1 mM PMSF (serine proteases)
- 10 μg/mL leupeptin (cysteine/threonine proteases)
- 1 μg/mL pepstatin A (aspartic proteases)
- 10 mM EDTA (metalloproteases)
Critical note: CTAB precipitates EDTA; use EGTA instead for cationic detergents.
Purification Challenges Specific to Ionic Detergents
Chromatography Incompatibilities: A Detailed Analysis
Ion-Exchange Chromatography (IEX)
The most impacted technique due to direct charge competition:
- Anionic detergents (SDS, DOC) compete with protein for binding to anion-exchange resins (Q-column), causing premature elution in flow-through
- Cationic detergents (CTAB) bind irreversibly to cation-exchange resins (S-column), contaminating the matrix
Practical workaround:
- Pre-clearing: Run detergent alone through column to saturate binding sites (wasteful but effective)
- Detergent reduction: Dilute sample 10-fold to reduce free detergent monomer concentration below CMC before loading
- Switch to alternative chromatography: Use IMAC or affinity tags instead of IEX
Size Exclusion Chromatography (SEC)
Micelle interference creates major artifacts:
- Apparent MW inflation: A 50 kDa membrane protein in SDS micelles elutes at ~130 kDa, misleading oligomeric state assessment
- Resolution loss: Large micelles reduce pore accessibility, broadening peaks
- Non-ideal behavior: Micelle-protein dissociation during SEC can cause on-column aggregation
Mitigation strategies:
- Column calibration: Use protein standards prepared in identical detergent conditions
- Detergent supplementation: Include 0.1-0.5×CMC detergent in running buffer to maintain PDC stability
- High-resolution SEC: Use columns with ≥30 cm bed height and small particle size (≤10 μm) to maximize separation
Affinity Chromatography
IMAC (Immobilized Metal Affinity Chromatography):
- Generally more tolerant than IEX; Ni-NTA resins can function in 0.1% SDS or 1% DOC
- Critical parameter: Imidazole concentration must be optimized independently as detergents can chelate Ni²⁺ weakly
Strep-Tactin/Strep-tag:
- Highly sensitive to SDS; >0.05% causes complete binding failure
- Solution: Buffer exchange to non-ionic detergent before loading
Molecular Mechanisms of Activity Loss
Transmembrane Domain Destabilization: Ionic detergents insert between transmembrane helices, disrupting hydrophobic packing. For 7-transmembrane GPCRs, SDS can reduce helical content by 30-40% as measured by circular dichroism.
Quaternary Structure Disruption: Many membrane proteins function as dimers or higher oligomers. SDS's strong negative charge can repel subunits, dissociating functional complexes. The mitochondrial ADP/ATP carrier loses dimeric structure in SDS, becoming monomeric and inactive.
Essential Lipid Stripping: Annular lipids (boundary lipids) often constitute integral components of membrane protein structure. Ionic detergents, being more "aggressive" than non-ionic, strip these lipids more efficiently, removing structural lipids like cardiolipin from respiratory complexes, leading to activity loss.
Mitigation Strategies: Preserving Activity During Ionic Detergent Extraction
Detergent-Lipid Mixed Micelles
Maintaining a "native-like lipid environment":
Add synthetic lipids at 0.1-0.2:1 lipid-to-detergent molar ratio:
- POPC (16:0-18:1 PC): For general eukaryotic membrane proteins
- POPE/POPG mixture (3:1): For bacterial inner membrane proteins
- Cardiolipin (CL): Essential for respiratory complexes and mitochondrial carriers
Mechanism: Lipids form a "halo" around the protein transmembrane region, partially shielding it from direct detergent contact. This maintains local lipid-protein interactions crucial for activity. Studies on the lactose permease (LacY) showed that adding 0.1% E. coli polar lipid extract during DDM solubilization retained 85% transport activity versus 30% without lipids.
Rapid Detergent Exchange Techniques
On-Column Exchange
IMAC workflow:
- Bind His-tagged protein in ionic detergent (e.g., 1% DOC)
- Wash extensively (10-15 column volumes) to remove unbound detergent
- Elute with high imidazole buffer containing target non-ionic detergent (0.05% DDM)
- Protein elutes in DDM, achieving >95% detergent exchange in a single step
Advantages: Minimizes time spent in harsh detergent, typically completing exchange within 2-3 hours.
Detergent Adsorption
For low-CMC detergents (SDS, CTAB) that resist dialysis:
- Bio-Beads SM-2: Hydrophobic polystyrene beads that adsorb detergent monomers
- Protocol: Mix sample with pre-washed Bio-Beads (100 mg/mL) at 4°C for 2-4 hours
- Efficiency: Removes >90% SDS in 4 hours without significant protein loss
- Caution: Rapid removal can cause aggregation; monitor by DLS
Stabilizer Synergism
Additive cocktails to counteract denaturation:
- Glycerol (10-20% v/v): Increases solution viscosity, reducing detergent diffusion and collisional denaturation
- Sucrose (0.5-1 M): Preferential hydration stabilizes native state
- L-Arginine (50-200 mM): Suppresses aggregation by binding to exposed hydrophobic patches
- Specific ligands: Substrate/inhibitor binding can lock proteins in stable conformations during solubilization
Example: The glutamate transporter GLT-1 retained 70% activity when solubilized in 1% DOC supplemented with 200 mM L-arginine and 10% glycerol, versus <20% without additives.
Function-Verification Node Design
Implement activity checks at each purification stage:
- Post-solubilization: Ligand binding assay (e.g., radiolabeled substrate)
- Post-IMAC: Enzymatic activity assay
- Post-SEC: Thermal shift assay (DSF) to assess conformational stability
Acceptable thresholds:
- Functional yield >50% of starting material
- Specific activity within 2-fold of native membrane preparation
- Monodisperse SEC profile with minimal aggregation peak
Comparative Analysis: Strategic Decision Framework
Workflow Decision Tree
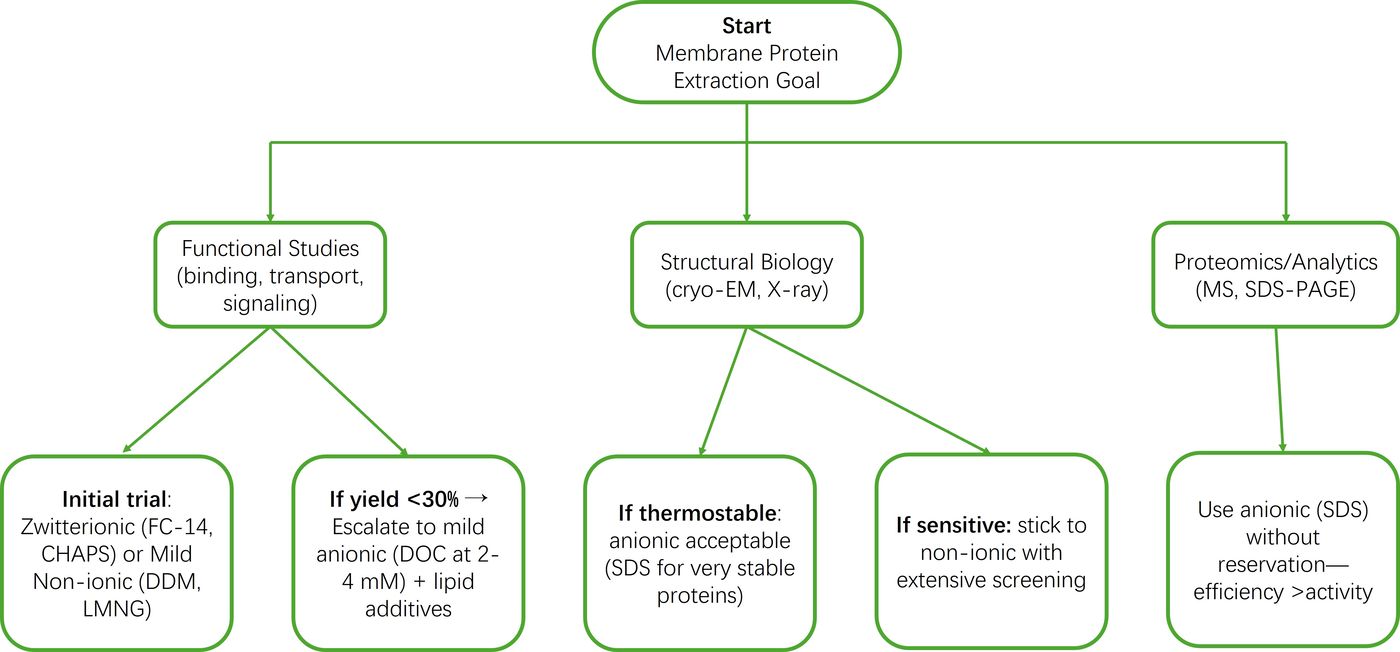
Guiding principle: Always start with the mildest detergent achieving >50% extraction in pilot experiments.
The Yield-Functionality Trade-off: Quantitative Analysis
Case study: Isolation of the human serotonin transporter (hSERT)
| Detergent | Extraction Yield | [³H]Imipramine Binding (Ki) | Oligomeric State | Functional Yield |
|---|---|---|---|---|
| DDM (1%) | 35% | 1.2 nM (native) | Dimer | 32% |
| DOC (4 mM) | 68% | 3.8 nM (3-fold loss) | Monomer-dimer mix | 45% |
| SDS (1%) | 95% | No binding | Monomer (denatured) | 0% |
This demonstrates DOC's strategic advantage: doubling extraction while retaining partial function, enabling downstream applications impossible with DDM alone.
Cost-Benefit Analysis
Economic considerations:
- SDS: $0.50/g, extremely cost-effective for large-scale membrane proteomics
- DOC: $5-10/g, moderate cost, requires careful optimization
- DDM: $50-100/g, expensive but necessary for high-value functional proteins
Time investment: DOC optimization requires 3-5 days of screening; DDM often works "out of the box." For precious samples (e.g., primary tissue, low-expression constructs), the time savings justify DDM's cost. For bacterial overexpression systems, DOC offers compelling economics.
Case Studies: Successful Applications in Detail
Case Study 1: Deoxycholate Extraction of YedZ-GFP-His
Background: YedZ, a heme-binding membrane protein from E. coli, expressed as GFP fusion with low solubility in DDM.
Protocol optimization:
- Membrane preparation: French press lysis, ultracentrifugation at 100,000×g
- Solubilization: 1% deoxycholate, 50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM PMSF
- Incubation: 30 min at 4°C with gentle rotation
- Clarification: 100,000×g ultracentrifugation for 45 min
- IMAC: Ni-NTA chromatography with 0.1% DOC in all buffers
- Detergent exchange: On-column exchange to 0.05% DDM during elution
- Polishing: SEC in DDM buffer
Outcome:
- Yield: 3 mg/L culture (3-fold higher than DDM)
- Activity: Heme content per protein ratio 0.85 (native membrane: 1.0)
- Purity: >95% monodisperse peak by SEC
- Key success factor: Low temperature + glycerol + rapid DOC-to-DDM exchange preserved heme binding pocket integrity
Case Study 2: CTAB Extraction of Acidic Membrane Proteins
Challenge: Purifying lactate dehydrogenase complex from mitochondrial inner membrane rich in cardiolipin (negative charge density).
CTAB strategy:
- Extraction: 0.5% CTAB, 20 mM HEPES pH 7.5, 50 mM NaCl (low salt prevents precipitation)
- Removal: Immediate post-extraction treatment with Bio-Beads SM-2 (150 mg/mL) for 4 hours at 4°C
- Re-solubilization: CTAB-depleted pellet re-dissolved in LMNG buffer for functional assays
Result: CTAB extracted 80% of protein versus 25% with DDM; Bio-Beads removed 92% CTAB; final reconstituted complex retained 65% enzymatic activity. This two-step approach leveraged CTAB's unique affinity for acidic membranes while mitigating its incompatibility through removal and reconstitution.
Case Study 3: SDS-Based Refolding of GPCR Inclusion Bodies
Background: Purified human adenosine A₂A receptor expressed in E. coli as inclusion bodies required complete denaturation and refolding.
Refolding protocol:
- Solubilization: 8 M guanidine-HCl + 2% SDS, 50°C for 1 hour
- Dilution: Rapid 100-fold dilution into refolding buffer (50 mM Tris pH 7.5, 0.1% SDS, 0.5 M arginine, 10% glycerol)
- Detergent exchange: Dialysis against 0.05% SDS → 0.01% SDS → 0.05% DDM over 72 hours
- Affinity purification: IMAC capture in DDM buffer
Success metrics:
- Refolding efficiency: ~15% (low but usable)
- Ligand binding: [³H]ZM241385 binding with Kd 8 nM (close to native 3 nM)
- Crystallization: Yielded diffraction-quality crystals
Lesson: SDS-based denaturation-refolding is viable for robust proteins but requires extensive optimization and yields are modest.
Emerging Trends and Next-Generation Solutions
Rational Detergent Design: HLB Optimization
Fluorinated analogs: Perfluorooctanoic acid (PFOA) and perfluorooctyl sulfonate (PFOS) demonstrate:
- High solubilization power: Comparable to SDS
- Reduced denaturation: Fluorocarbon tails are more rigid, creating ordered micelles
- Chemical stability: Resistant to oxidation
Custom HLB tuning: By varying alkyl chain length (C10-C16) and head group charge density, researchers can fine-tune detergents for specific protein classes. Machine learning models now predict optimal HLB values based on transmembrane domain hydrophobicity and oligomeric state.
Detergent-Free Technologies: The SMALP Revolution
Styrene-maleic acid lipid particles (SMALP) offer a paradigm shift:
- Mechanism: SMA polymer inserts into membranes, directly extracting disk-like lipid bilayers containing embedded proteins—no detergent required
- Advantages: Native lipid environment, no detergent artifacts, superior activity retention
- Limitations: Requires optimization of polymer:protein ratio; not compatible with all buffer conditions
Direct comparison: SMALP extraction of the ABC transporter BtuCD retained 98% ATPase activity versus 55% with DOC and 0% with SDS.
Hybrid Systems: Synergistic Ionic/Non-Ionic Mixtures
Strategic combinations:
- 0.1% SDS + 0.5% DDM: SDS disrupts tight lipid packing, DDM stabilizes protein
- 2 mM DOC + 1% glyco-diosgenin (GDN): DOC boosts extraction, GDN preserves oligomeric state
Synergy mechanism: Components partition into different micelle regions—ionic head groups at protein surface, non-ionic tails forming bilayer-like environment. This cooperative effect can increase functional yield by 1.5-2 fold compared to single detergents.
AI-Driven Detergent Screening
Machine learning models (e.g., DeepSol, MPNext) trained on thousands of detergent-protein combinations now predict:
- Optimal detergent class based on amino acid composition and topology
- Expected extraction yield and functional retention
- Compatibility with downstream applications
Validation: AI-predicted optimal detergents matched experimental best performers in 78% of cases, reducing screening time from weeks to days.
Advanced Diagnostics
For persistent issues, implement:
- Thermal shift assay: Compare Tm values in different detergents to assess stability
- Analytical ultracentrifugation (AUC): Determine true oligomeric state independent of micelle effects
- Mass photometry: Single-molecule assessment of aggregation state
- Ligand binding with FRET: Sensitive probe of conformational integrity
Conclusion: Navigating the Ionic Detergent Landscape
Ionic detergents, though historically viewed as "last resort" agents, possess unique capabilities that, when harnessed strategically, can unlock recalcitrant membrane proteins inaccessible to gentler methods. The future of membrane protein isolation lies not in detergent elimination, but in intelligent application:
- Embrace the MEC principle: Start at 1×CMC, titrate upward only as needed
- Multiparameter optimization: Simultaneously adjust detergent, lipid additives, temperature, and buffer conditions
- Rapid processing: Minimize time between solubilization and stabilization in mild detergent
- Functional validation: Prioritize activity metrics over sheer extraction yield
- Hybrid approaches: Combine ionic extraction power with non-ionic stabilization through exchange
Emerging paradigms promise to transform the field. AI-driven screening will predict optimal conditions in silico, reducing empirical burden. Rational detergent design will yield "smart" ionic detergents with tunable harshness. Most radically, detergent-free technologies like SMALP may eventually supersede traditional methods for functional studies, preserving native lipid environments unparalleled by any artificial micelle system.
Final recommendation: For functional membrane protein research, never default to ionic detergents. Begin with gold-standard non-ionic agents. If yield is insufficient after exhaustive optimization, systematically transition through zwitterionic, then mild ionic (DOC), reserving harsh detergents (SDS) for analytical or structural applications where activity is secondary. This tiered, evidence-based approach ensures maximal biological relevance while capturing the full membrane proteome spectrum.
Related Products & Services
Resources
-

Enhance Your Biomedical Research With Our High Quality Transmembrane Proteins
-
Transmembrane Protein
-
Mastering Membrane Protein Purification: The Detergent Dilemma
-
Picking the Perfect Detergent for Membrane Proteins
-
Master Detergent Selection in 60 Seconds
References
- Methods in Enzymology, Vol. 557, "Membrane Proteins: Expression, Purification, and Characterization" (2015)
- B. K. Kobilka, "GPCR Structure and Activation: A Membrane Protein Perspective" Nature Reviews Mol Cell Biol 2020
- M. J. le Maire et al., "Detergents as Tools in Membrane Biochemistry" J. Biol. Chem. 2019
- S. Schlegel et al., "Bacterial-Based Membrane Protein Production" Biochim. Biophys. Acta 2021
- J. L. Parker et al., "Strategies for Solubilizing Membrane Proteins" Methods 2022
- A. D. Duell et al., "Ionic Detergents in Membrane Protein Purification" Protein Science 2023
- T. H. Bayburt et al., "Detergent Optimization for Membrane Proteins" Anal. Biochem. 2020
- T. J. Knowles et al., "Membrane Protein Stability in Detergent Solutions" Biochemistry 2021
- R. B. Booth et al., "Rational Design of Novel Detergents for Membrane Proteins" JACS 2022
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

