Protocol: Detergent Selection and Optimization for Membrane Protein Purification
Pre-Experimental Planning: Strategic Detergent Selection
Initial Detergent Choice Based on Target Protein Characteristics
Step 1: Define Your Priorities
Before touching any reagent, establish your non-negotiable requirements:
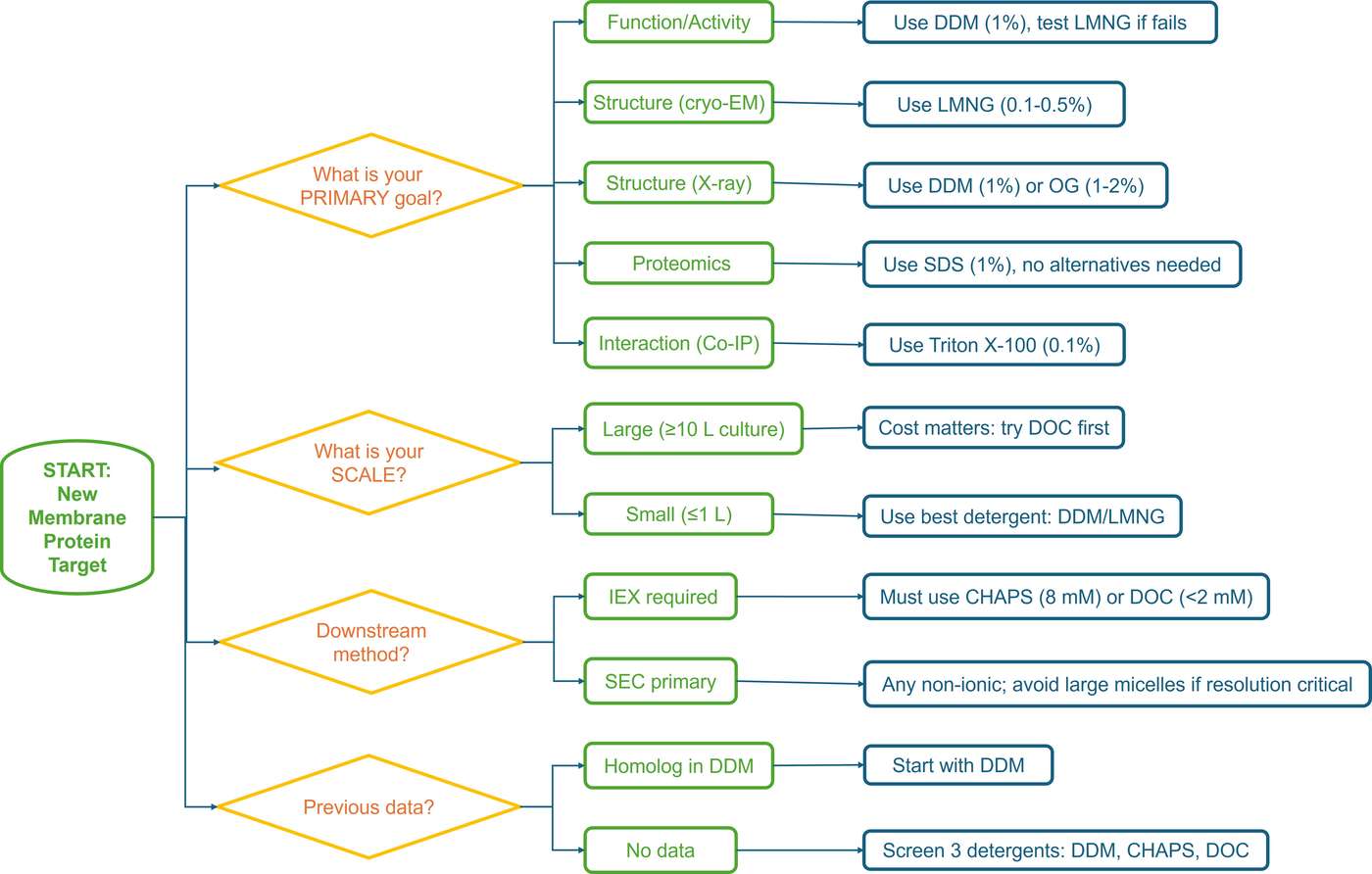
- If functional activity is required (ligand binding, enzymatic assays, transport): Primary candidates: DDM (1%), LMNG (0.1%), or GDN (0.1%) Avoid: All ionic detergents (SDS, CTAB) as first choice
- If structural determination is the goal (cryo-EM, crystallography): Primary candidates: LMNG (0.1-0.5%) for cryo-EM; DDM (1%) or OG (1-2%) for crystallography Consider: Add CHS (0.1-0.2%) for GPCRs
- If proteomic coverage is the priority: Primary candidate: SDS (1-2%) for complete denaturation and extraction Accept the trade-off: Functional loss is inevitable
- If ion-exchange chromatography is essential: Primary candidates: CHAPS (8 mM) or low-concentration DOC (2 mM) Avoid: SDS, CTAB, high-concentration ionic detergents
Step 2: Literature Mining
Search PubMed for your specific protein or homologs. Extract:
- Detergent types used (focus on most recent papers)
- Concentrations reported (note both solubilization and purification concentrations)
- Additives mentioned (lipids, glycerol, ligands)
- Yield and activity data if available
Step 3: Calculate Your CMC Budget
Determine the minimum detergent needed:
- Total membrane protein: 5-10 mg/mL in your solubilization buffer
- Target detergent concentration: 1-3×CMC (start with 2×CMC)
- Volume: Plan for 10-20% excess to account for losses
Example: For DDM (CMC = 0.17 mM = 0.0086% w/v), 2×CMC = 0.017% w/v. For 10 mL solubilization, you need ~1.7 mg DDM. A 1 g bottle costs ~$75, so reagent cost per experiment is negligible, but optimization costs add up.
Buffer Design & Additive Optimization
Master Buffer Formulation
Base Buffer (for all conditions):
- Buffering agent: 20-50 mM HEPES or Tris, pH 7.5-8.0 (match detergent pKa)
- Salt: 150 mM NaCl (standard) or 100-500 mM range for optimization
- Stabilizers: 10-20% glycerol (v/v), 5-10% sucrose (w/v)
- Reducing agent: 1-5 mM TCEP (preferred over DTT; more stable, no odor)
- Add last: Protease inhibitor cocktail (fresh)
Protease Inhibitor Cocktail (add immediately before solubilization):
- 1 mM PMSF (or 0.1 mM AEBSF, safer)
- 10 μg/mL leupeptin
- 1 μg/mL pepstatin A
- 5 mM EDTA (or EGTA if using CTAB)
Critical Note: CTAB + EDTA = precipitate. Always use EGTA with cationic detergents.
Lipid Supplementation (Essential for Functional Work)
When to add: Always for GPCRs, transporters, and complexes known to require lipids
Standard lipid mix (0.1-0.2:1 lipid-to-detergent molar ratio):
- Eukaryotic proteins: POPC (16:0-18:1 PC) + POPE (16:0-18:1 PE) at 3:1 ratio
- Bacterial proteins: POPE:POPG (3:1) mix
- Respiratory complexes: Add 10% cardiolipin (CL)
- GPCRs: Add CHS (cholesteryl hemisuccinate) at 0.1-0.2%
Preparation: Prepare 10× lipid stock in detergent-containing buffer by sonication until clear. Add to solubilization mixture.
Additional Stabilizers
For recalcitrant proteins (add to base buffer):
- Osmolyte: 50-200 mM L-argininine (reduces aggregation)
- Salting-in: 0.5-1 M NaCl (can improve solubilization)
- Chelator: 5 mM EDTA (prevents metal-catalyzed oxidation)
- Substrate/inhibitor: If available, add at saturating concentration
Step-by-Step Solubilization Protocol
Pre-Solubilization Checklist
□ Membrane pellets resuspended to 5-10 mg/mL total protein
□ Detergent stocks prepared at 10× final concentration
□ All buffers chilled to 4°C (or optimized temperature)
□ Protease inhibitors freshly added
□ Lipid stocks prepared and warmed if necessary
□ Control samples prepared (no detergent, detergent-only)
Controlled Solubilization Workflow
Time: 30-60 minutes | Temperature: 4°C (standard) or optimized
- Aliquot membrane suspension: Dispense into ultracentrifuge tubes (e.g., 1 mL per tube for small-scale)
- Add detergent:
- Slowly pipette detergent stock while vortexing gently
- Final concentration: 1-3×CMC (start with 2×CMC)
- Example: For DDM at 2×CMC, add 0.017% w/v final
- Incubate:
- Place on rotating mixer or nutator at 4°C
- Time: 30 minutes (standard) to 60 minutes (for stubborn membranes)
- Critical: Do not exceed 60 min for functional work; set timer
- Monitor viscosity: If solution becomes extremely viscous, you've reached the "gel phase"—dilute immediately with 1× buffer
- Clarification:
- Transfer to ultracentrifuge tubes (ensure balanced)
- Spin at 100,000×g for 45 minutes at 4°C
- Carefully remove supernatant without disturbing lipid layer at top or pellet
- Assess extraction:
- Take 20 μL supernatant and 20 μL pre-spin sample
- Run SDS-PAGE and stain with Coomassie or silver
- Target: >50% target protein in supernatant
Time-Course Optimization (If Extraction is Poor)
If initial extraction <30%, perform time-course:
| Time Point | Duration | Temperature | Sample Collection |
|---|---|---|---|
| T₁ | 15 min | 4°C | Spin separate tube |
| T₂ | 30 min | 4°C | Spin separate tube |
| T₃ | 60 min | 4°C | Spin separate tube |
| T₄ | 30 min | RT (22°C) | Compare to 4°C |
Analyze: Plot % extraction vs time. Stop at plateau to minimize denaturation.
Detergent Removal & Exchange Protocols
Method Selection Based on CMC
Flowchart: Which Method to Use?
Q: What is your detergent's CMC?
1) >5 mM (OG, CHAPS, DOC) → Use DIALYSIS or DILUTION → Protocol 1
2) 1-5 mM → Use DIALYSIS (slow) or ADSORPTION (fast) → Protocol 2
3) <1 mM (DDM, LMNG, SDS, CTAB) → Use ADSORPTION or ON-COLUMN EXCHANGE → Protocol 2/3
Protocol 1: Dialysis (High CMC Detergents)
Equipment: SnakeSkin dialysis tubing (3.5-10 kDa MWCO) or Float-A-Lyzer
Steps:
- Dilute sample 1:1 with detergent-free buffer (reduces detergent concentration by 50%)
- Load into dialysis tubing (leave 30% headspace for expansion)
- Dialyze against 100× volume of buffer at 4°C
- Buffer changes: Every 6-8 hours for 48-72 hours minimum
- Verification: Test effluent with CMC assay (surface tension) or mass spec
Expected removal: >95% for OG, ~80% for DOC (due to moderate CMC)
Protocol 2: Hydrophobic Adsorption (Low CMC Detergents)
Equipment: Bio-Beads SM-2 (Bio-Rad) or Amberlite XAD-16
Preparation:
- Wash beads: 3× with methanol, 3× with deionized water, 3× with buffer
- Never use dry beads directly—they will adsorb protein!
Steps:
- Add washed Bio-Beads to sample: 100 mg beads per mL sample
- Incubate at 4°C with gentle rotation (not vortexing)
- Timeline: 2-4 hours (check at 2h and 4h)
- Remove beads by filtration or brief centrifugation (500×g)
- Check protein concentration (some loss due to non-specific adsorption is normal)
For SDS removal: May need 2-3 sequential treatments (replace beads each time)
Protocol 3: On-Column Detergent Exchange (Most Efficient)
Applied to: IMAC, Strep-Tactin, or other affinity chromatography
Workflow:
- Binding: Load solubilized protein onto column in "harsh" detergent (e.g., 1% DOC)
- Washing: Wash with 15-20 column volumes (CV) of binding buffer containing 0.1-0.5×CMC detergent
- Key: This removes bulk detergent micelles while keeping protein solubilized
- Exchange: Apply 5 CV of buffer containing target mild detergent (e.g., 0.05% DDM)
- Elution: Elute protein with buffer containing target detergent + elution agent (imidazole, biotin, etc.)
Result: >95% detergent exchange, minimal time in harsh conditions (<3 hours total)
Pro Tip: For His-tagged proteins, add 10 mM imidazole to all wash buffers to reduce non-specific binding
Protocol 4: Detergent Spin Columns (Rapid, Small Scale)
Commercial kits
Steps:
- Equilibrate column with 2 mL detergent-free buffer (centrifuge 500×g, 1 min)
- Load 0.5 mL sample
- Centrifuge 500×g for 2 minutes
- Collect flow-through (protein) and discard column (detergent)
Limitations:
- Capacity: Only removes free detergent, not protein-bound detergent
- Volume: Small scale only
- Efficiency: 70-90% removal
Integration with Purification Chromatography
SEC (Size Exclusion Chromatography) Optimization
Column preparation:
- Equilibrate with 2 CV of buffer containing 0.1-0.5×CMC detergent (never detergent-free!)
- Example: For DDM (CMC 0.17 mM), use 0.05 mM in running buffer
Calibration:
- Run standards prepared in identical detergent buffer
- Apparent MW inflation: Plan for +60-100 kDa for DDM, +18 kDa for SDS
Sample loading:
- Concentrate to 1-5 mg/mL (too high → aggregation; too low → poor resolution)
- Spin at 14,000×g for 10 min before loading to remove particulates
Peak interpretation:
- Monomer: Should be symmetric, baseline-resolved
- Aggregates: Void volume peak (V₀) or shoulders
- Lipid micelles: Separate peak at apparent MW 40-60 kDa
Troubleshooting SEC: See Section below
Ion-Exchange (IEX) Strategies
Compatible detergents: CHAPS, low DOC (<2 mM), amphipols
Incompatible: SDS, high DOC, CTAB
If you must use IEX with ionic detergent:
- Dilute sample 10× with detergent-free buffer (reduces free detergent concentration)
- Load immediately (protein may start to aggregate)
- Use shallow gradients (0-300 mM NaCl over 20 CV)
- Collect peaks quickly and supplement with stabilizing detergent
Better alternative: Perform IEX before solubilization (on membrane fraction) if possible
IMAC (His-Tag Purification) Specifics
Detergent compatibility: Generally tolerant, but optimize per detergent:
| Detergent | Max [ ] in binding buffer | Effect on binding |
|---|---|---|
| DDM | 0.1-0.5% | Minimal |
| SDS | 0.01-0.05% | Reduces by ~30% |
| DOC | 1% | Minimal |
| CHAPS | 2% | Minimal |
Critical: Add 10-20 mM imidazole to binding and wash buffers to reduce non-specific binding without affecting target protein binding
Detergent exchange: Use Protocol 4.3 (on-column exchange) for best results
Comprehensive Troubleshooting Decision Tree
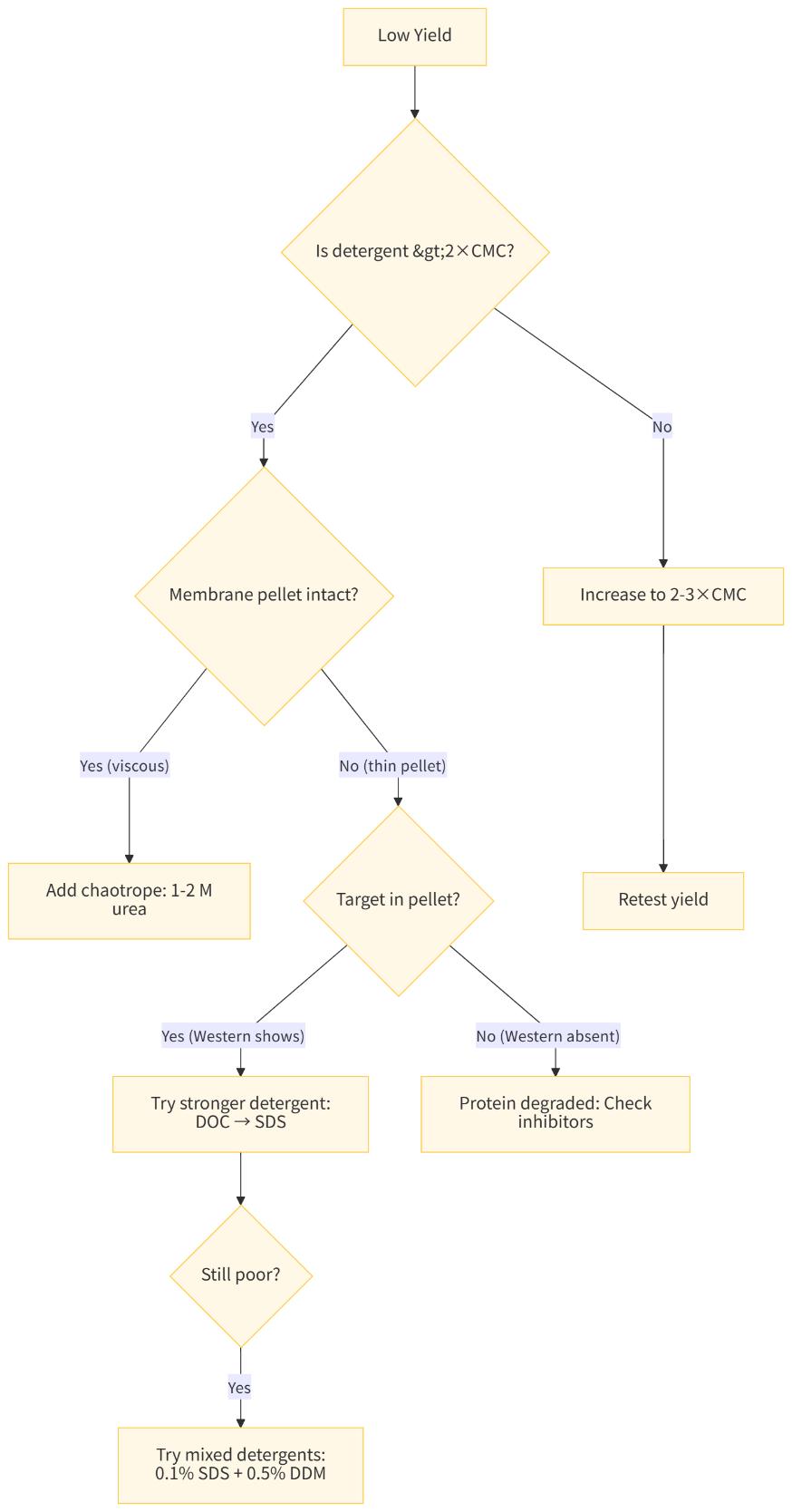
Problem: Low Extraction Yield (<30%)
Diagnostic Flow:
Action Steps:
- Verify CMC: Remeasure detergent stock concentration (some degrade)
- Increase concentration: Stepwise to 3×, 5×CMC (max for functional work)
- Add chaotrope: 1-2 M urea or 0.5-1 M NaCl (increases membrane fluidity)
- Try mixed system: 0.1% SDS + 0.5% DDM (synergistic extraction)
- Check membrane prep: Ensure membranes are not over-washed (loses peripheral proteins)
- Sonication: Brief probe sonication (3×5 sec) before detergent addition
Problem: Protein Fails to Bind Chromatography Column
For IMAC:
| Symptom | Cause | Test | Solution |
|---|---|---|---|
| Flow-through has target | Tag stripped by detergent | Western for tag | Reduce detergent or add 10 mM imidazole during solubilization |
| Low binding | Charge interference | Measure sample conductivity | Dilute 10× or add 100 mM NaCl to screen |
| No binding | His-tag inaccessible | Add 5 mM EDTA → remove | Try different tag location (C-term vs N-term) |
| Binds but elutes early | Weak binding | Check imidazole in wash | Increase imidazole gradient (0-500 mM) |
For IEX:
- If using ionic detergent: Must be zwitterionic or non-ionic. Switch immediately.
- If using CHAPS/DOC: Dilute sample 5× before loading to reduce micelle interference
Problem: Heavy Aggregation (SEC shows void peak)
Diagnostic Tests:
- DLS of supernatant: Size >100 nm = aggregation
- SDS-PAGE of pellet: Insoluble material = misfolded aggregates
- Native PAGE: High MW smears = oligomeric aggregates
Solution Matrix:
- If aggregation in all fractions: Reduce protein concentration (dilute 5×), add 200 mM arginine
- If aggregation only in concentrated peaks: Concentrate more slowly, add 20% glycerol
- If aggregation with age: Fresh prep, reduce purification time, add 5 mM DTT
- If aggregation at 4°C: Try RT (22°C) but reduce time to 10 min
Advanced: Cross-linking analysis (BS3) to determine if aggregates are covalent
Problem: Multiple SEC Peaks / Poor Resolution
Interpretation Guide:
| Peak Position | Possible Identity | Verification | Action |
|---|---|---|---|
| Void volume (V₀) | Aggregates | DLS | Add stabilizers, reduce concentration |
| 1.5× expected MW | Detergent micelle only | No protein in peak | Increase wash to remove free micelles |
| Broad shoulder | Heterogeneous oligomers | Native PAGE | Add lipid, optimize detergent |
| Split main peak | Conformational heterogeneity | Thermal shift | Add ligand to lock conformation |
Optimization:
- Column: Use longer column (≥60 cm) or smaller bead size
- Flow rate: Reduce to 0.3-0.4 mL/min for better resolution
- Sample volume: Max 2% of column volume (e.g., 0.5 mL for 24 mL column)
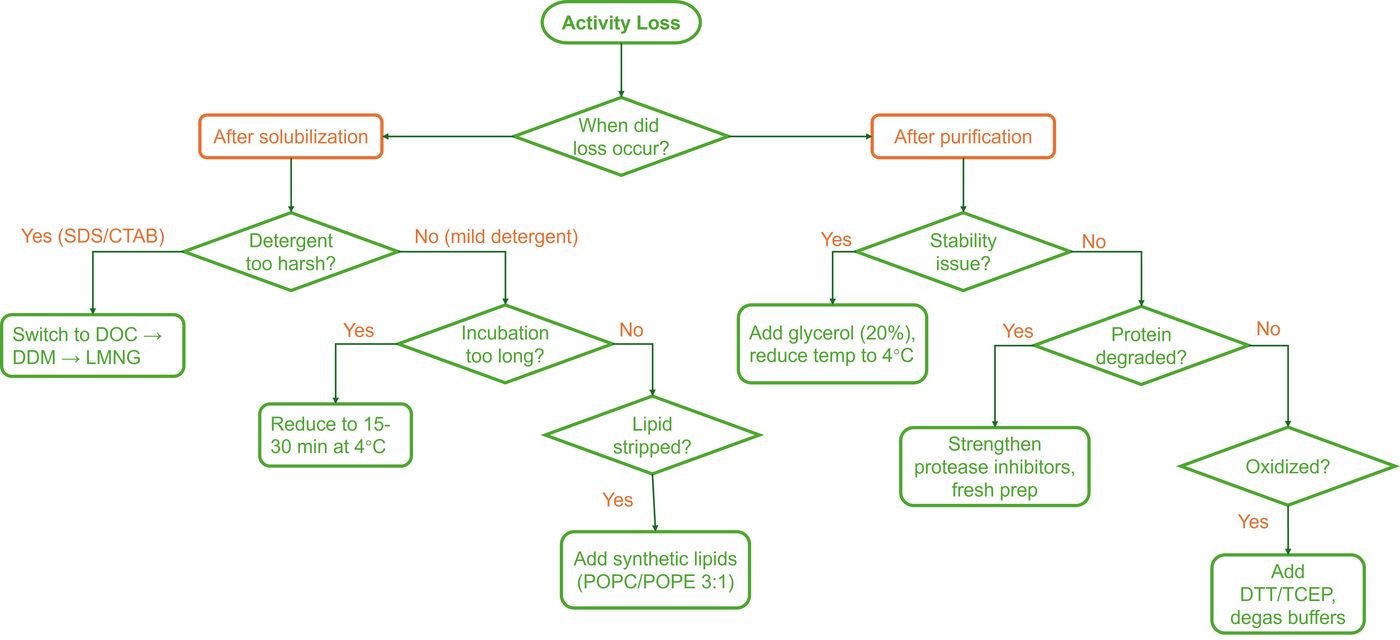
Problem: Activity Loss >50%
Systematic Diagnosis:
Functional Assay Controls:
- Solubilization control: Assay membranes before solubilization (100% activity baseline)
- Detergent control: Assay protein after buffer exchange to non-ionic
- Recovery control: Assay SEC peak fractions separately to identify where activity lost
Critical Threshold: If activity retained <30% at any step, STOP and troubleshoot before scaling up.
Problem: Detergent-Specific Issues
SDS Precipitation:
- Cause: High K⁺, low temperature, old stock (>6 months)
- Solution: Use Na⁺-based buffers only; prepare fresh SDS monthly; if precipitates form, warm to 37°C
CTAB Precipitation:
- Cause: High salt, EDTA, freeze-thaw
- Solution: Keep NaCl <50 mM; use EGTA not EDTA; never freeze; maintain at 4°C consistently
Triton X-100 Clouding:
- Cause: Above cloud point (~65°C) or in presence of PEG
- Solution: Work at RT or 4°C; avoid PEG precipitants; if clouded, chill on ice and spin
DDM/LMNG Micelle Crystals:
- Cause: High concentration + low temp
- Solution: Warm to 30°C for 10 min; filter through 0.22 μm; store at RT
Quick Reference Tables
Detergent Property & Application Matrix
| Detergent | CMC (mM) | CMC (% w/v) | Strongest Suitability | Key Limitation | Removal Method |
|---|---|---|---|---|---|
| SDS | 8.2 | 0.24 | Proteomics, analytics | Irreversible denaturation | Bio-Beads (difficult) |
| DOC | 5.0 | 0.21 | Functional extraction | Can denature, pH-sensitive | Dialysis (moderate) |
| DDM | 0.17 | 0.0086 | Gold standard functional | Expensive, low CMC | Bio-Beads, hard |
| LMNG | 0.01 | 0.0006 | Cryo-EM, fragile proteins | Very expensive, ultra-low CMC | Bio-Beads, very hard |
| CHAPS | 6-8 | 0.4-0.5 | IEX-compatible | Moderate harshness | Dialysis (easy) |
| OG | 25 | 0.7 | Rapid removal | Harsh, poor stability | Dialysis (very easy) |
| CTAB | 0.9 | 0.033 | Acidic membranes | Very hard to remove, toxic | Bio-Beads (very hard) |
| Triton X-100 | 0.2-0.9 | 0.01-0.06 | Co-IP, mild extraction | UV interference, heterogeneous | Adsorption (hard) |
Troubleshooting Quick-Reference
| Problem | First Action | Second Action | Third Action |
|---|---|---|---|
| Low yield | Increase to 3×CMC | Add 1 M NaCl | Try mixed detergents |
| No IEX binding | Dilute 10× | Switch to CHAPS | Try affinity tag instead |
| SEC aggregates | Add 200 mM Arg | Reduce conc. 5× | Add 20% glycerol |
| Activity lost | Reduce time to 15 min | Add lipids | Switch to LMNG |
| CTAB precipitates | Dilute to <50 mM NaCl | Replace EDTA with EGTA | Use fresh, never freeze |
| SDS precipitates | Warm to 37°C | Filter 0.22 μm | Use Na⁺-only buffers |
| High background | Add 10 mM imidazole | Increase wash to 15 CV | Re-equilibrate column |
Decision Tree for Detergent Selection
Final Validation Checklist
Before declaring success, verify:
- Yield: >1 mg/L culture (or >50% recovery from starting material)
- Purity: >95% by SDS-PAGE densitometry
- Monodispersity: Single symmetric peak in SEC with <15% polydispersity by DLS
- Activity: ≥70% of native membrane activity retained
- Detergent residual: <0.01% if removed (mass spec or CMC assay)
- Stability: Stable at 4°C for 1 week (no aggregation)
- Concentration: Can concentrate to >5 mg/mL without precipitation
If all criteria met, proceed to functional/structural studies. If ≥2 criteria fail, repeat optimization with alternative detergent.
Related Products & Services
Resources
-

Enhance Your Biomedical Research With Our High Quality Transmembrane Proteins
-
Transmembrane Protein
-
Mastering Membrane Protein Purification: The Detergent Dilemma
-
Picking the Perfect Detergent for Membrane Proteins
-
Master Detergent Selection in 60 Seconds
Contact us or send an email at for project quotations and more detailed information.
Quick Links
-

Papers’ PMID to Obtain Coupon
Submit Now -

Refer Friends & New Lab Start-up Promotions

