Recombinant Rat MATR3 Protein
| Cat.No. : | MATR3-3598R |
| Product Overview : | Recombinant Rat MATR3 full length or partial length protein was expressed. |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Rat |
| Source : | Mammalian Cells |
| Tag : | His |
| Form : | Liquid or lyophilized powder |
| Endotoxin : | < 1.0 EU per μg of the protein as determined by the LAL method. |
| Purity : | >80% |
| Notes : | This item requires custom production and lead time is between 5-9 weeks. We can custom produce according to your specifications. |
| Storage : | Store it at +4 ºC for short term. For long term storage, store it at -20 ºC~-80 ºC. |
| Storage Buffer : | PBS buffer |
| Gene Name | Matr3 matrin 3 [ Rattus norvegicus ] |
| Official Symbol | MATR3 |
| Gene ID | 29150 |
| mRNA Refseq | NM_019149.3 |
| Protein Refseq | NP_062022.2 |
| MIM | |
| UniProt ID | P43244 |
| ◆ Recombinant Proteins | ||
| MATR3-4881HFL | Recombinant Full Length Human MATR3 protein, Flag-tagged | +Inquiry |
| MATR3-2688R | Recombinant Rhesus monkey MATR3 Protein, His-tagged | +Inquiry |
| MATR3-3598R | Recombinant Rat MATR3 Protein | +Inquiry |
| Matr3-3973M | Recombinant Mouse Matr3 Protein, Myc/DDK-tagged | +Inquiry |
| MATR3-3254R | Recombinant Rat MATR3 Protein, His (Fc)-Avi-tagged | +Inquiry |
| ◆ Cell & Tissue Lysates | ||
| MATR3-4447HCL | Recombinant Human MATR3 293 Cell Lysate | +Inquiry |
| MATR3-371HKCL | Human MATR3 Knockdown Cell Lysate | +Inquiry |
Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3
Journal: eLife PubMed ID: 28580901 Data: 2017/6/5
Authors: Ritu Chaudhary, Berkley Gryder, Joaquín M Espinosa
Article Snippet:Matrin 3 bound RNAs were isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation and qRT-PCR was used to determine the enrichment of p21 (negative control) and PINCR in the Matrin 3 IPs.Matrin 3 bound RNAs were isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation and qRT-PCR was used to determine the enrichment of p21 (negative control) and PINCR in the Matrin 3 IPs.. To determine the direct binding of PINCR to recombinant Matrin 3 (rMatrin 3), 200 ng of in vitro transcribed Bi- PINCR or Bi- LUC RNA was incubated with 500 ng recombinant Matrin 3 protein (Creative BioMart, Catalog # MATR3-15H) in 1X EMSA buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 0.1% Triton-X-100, 100 μg/ml BSA, 2 mM DTT and 5% glycerol) at room temperature for 2 hr.. RNA–protein complex was immunoprecipitated at room temperature for 2 hr, by using Dynabeads M-280 Streptavidin.RNA–protein complex was immunoprecipitated at room temperature for 2 hr, by using Dynabeads M-280 Streptavidin.
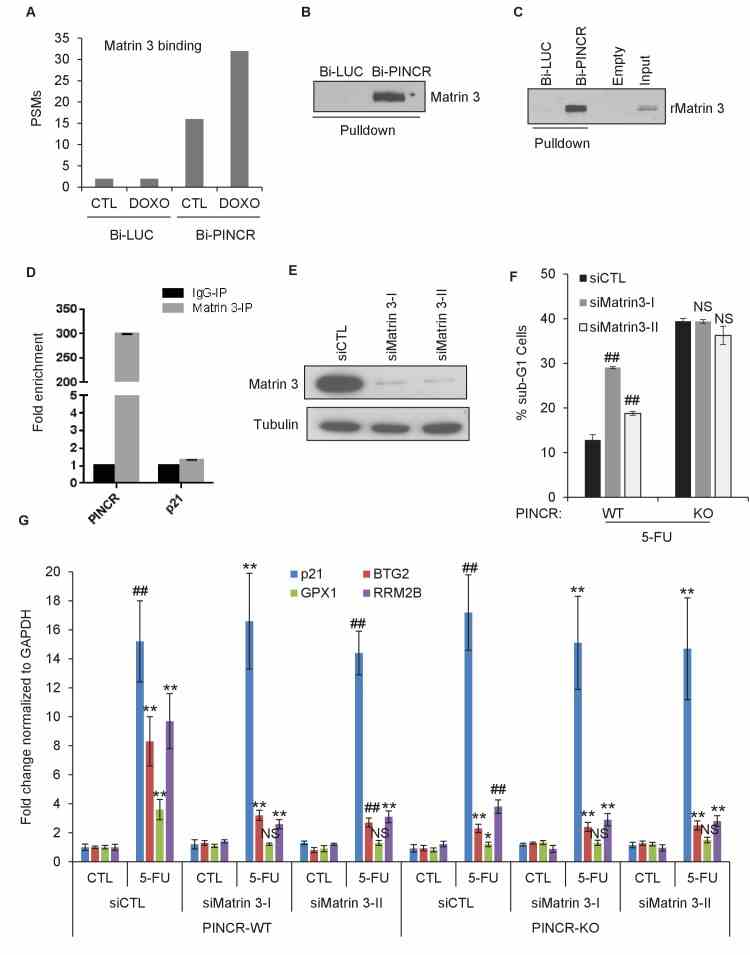
( A ) Peptide spectrum matches (PSMs) corresponding to
Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3
Journal: eLife PubMed ID: 28580901 Data: 2017/6/5
Authors: Ritu Chaudhary, Berkley Gryder, Joaquín M Espinosa
Article Snippet:Matrin 3 bound RNAs were isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation and qRT-PCR was used to determine the enrichment of p21 (negative control) and PINCR in the Matrin 3 IPs.Matrin 3 bound RNAs were isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation and qRT-PCR was used to determine the enrichment of p21 (negative control) and PINCR in the Matrin 3 IPs.. To determine the direct binding of PINCR to recombinant Matrin 3 (rMatrin 3), 200 ng of in vitro transcribed Bi- PINCR or Bi- LUC RNA was incubated with 500 ng recombinant Matrin 3 protein (Creative BioMart, Catalog # MATR3-15H) in 1X EMSA buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 0.1% Triton-X-100, 100 μg/ml BSA, 2 mM DTT and 5% glycerol) at room temperature for 2 hr.. RNA–protein complex was immunoprecipitated at room temperature for 2 hr, by using Dynabeads M-280 Streptavidin.RNA–protein complex was immunoprecipitated at room temperature for 2 hr, by using Dynabeads M-280 Streptavidin.
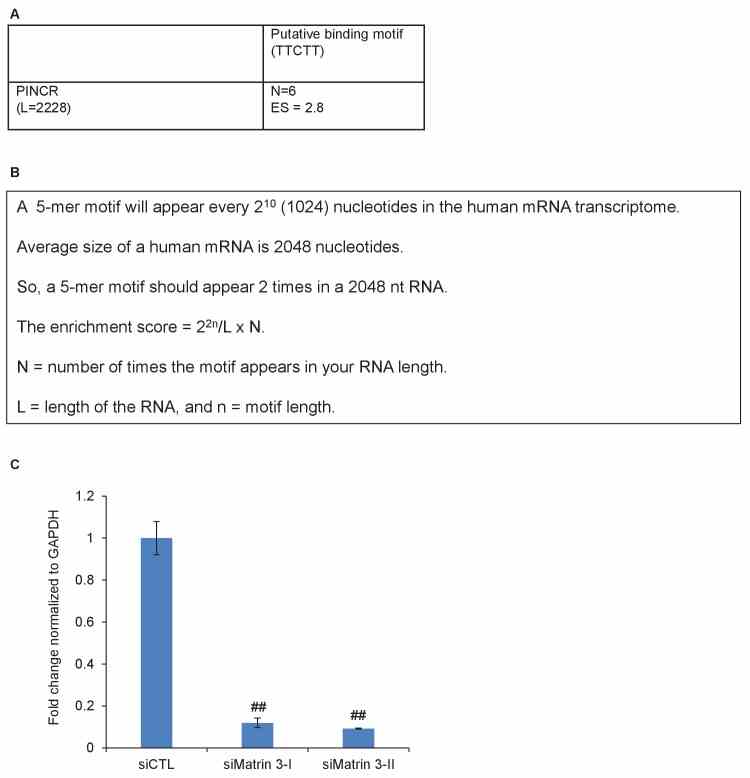
( A, B ) Putative
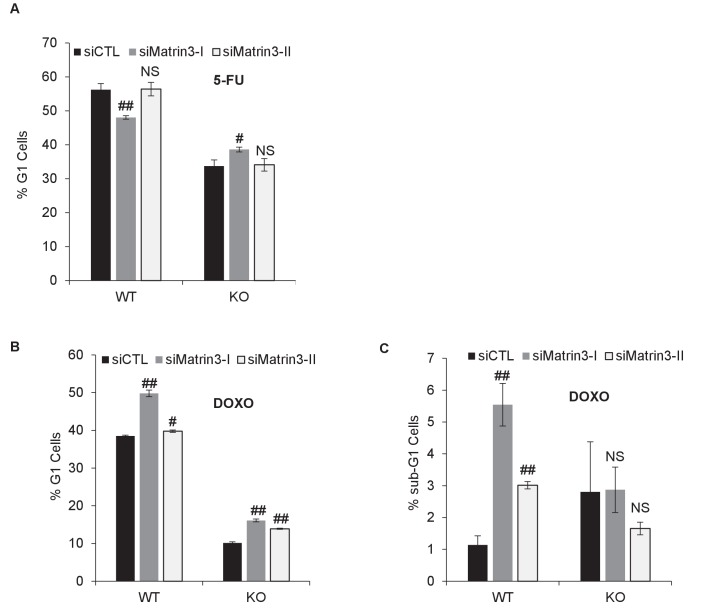
PINCR -WT and PINCR -KO HCT116 cells were reverse transfected with a control siRNA (siCTL) or siRNAs targeting
Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3
Journal: eLife PubMed ID: 28580901 Data: 2017/6/5
Authors: Ritu Chaudhary, Berkley Gryder, Joaquín M Espinosa
Article Snippet:Matrin 3 bound RNAs were isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation and qRT-PCR was used to determine the enrichment of p21 (negative control) and PINCR in the Matrin 3 IPs.Matrin 3 bound RNAs were isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation and qRT-PCR was used to determine the enrichment of p21 (negative control) and PINCR in the Matrin 3 IPs.. To determine the direct binding of PINCR to recombinant Matrin 3 (rMatrin 3), 200 ng of in vitro transcribed Bi- PINCR or Bi- LUC RNA was incubated with 500 ng recombinant Matrin 3 protein (Creative BioMart, Catalog # MATR3-15H) in 1X EMSA buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 0.1% Triton-X-100, 100 μg/ml BSA, 2 mM DTT and 5% glycerol) at room temperature for 2 hr.. RNA–protein complex was immunoprecipitated at room temperature for 2 hr, by using Dynabeads M-280 Streptavidin.RNA–protein complex was immunoprecipitated at room temperature for 2 hr, by using Dynabeads M-280 Streptavidin.

( A ) Peptide spectrum matches (PSMs) corresponding to

( A, B ) Putative

PINCR -WT and PINCR -KO HCT116 cells were reverse transfected with a control siRNA (siCTL) or siRNAs targeting
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All MATR3 Products
Required fields are marked with *
My Review for All MATR3 Products
Required fields are marked with *



