Recombinant Human SUMO1 Activating Enzyme Subunit 1, His-tagged
| Cat.No. : | SAE1-59H |
| Product Overview : | Recombinant human SAE1 protein, fused to His-T7-tag at N-terminus, was expressed inE.coliand purified by using conventional chromatography techniques. |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Human |
| Source : | E.coli |
| Tag : | His |
| Description : | SAE1, also known as AOS1, belongs to the ubiquitin-activating E1 family of proteins and plays an important role in the first step of the UBL1 conjugation pathway. Proteins conjugated to Ub are marked for progressive degradation by the 26S Proteasome. SAE1, which is a dimeric enzyme, functions as a UBLI E1 ligase mediating the ATP-dependent activation of UBL1. This protein can bind with UBLE1A and UBLE1B to form a heterodimer which can bind UBL1. |
| Sequences : | MHHHHHHMAS MTGGQQMGRD LYDDDDKDRW GSMVEKEEAG GGISEEEAAQ YDRQIRLWGL EAQKRLRASR VLLVGLKGLG AEIAKNLILA GVKGLTMLDH EQVTPEDPGA QFLIRTGSVG RNRAEASLER AQNLNPMVDV KVDTEDIEKK PESFFTQFDA VCLTCCSRDV IVKVDQICHK NSIKFFTGDV FGYHGYTFAN LGEHEFVEEK TKVAKVSQGV EDGPDTKRAK LDSSETTMVK KKVVFCPVKE ALEVDWSSEK AKAALKRTTS DYFLLQVLLK FRTDKGRDPS SDTYEEDSEL LLQIRNDVLD SLGISPDLLP EDFVRYCFSE MAPVCAVVGG ILAQEIVKAL SQRDPPHNNF FFFDGMKGNG IVECLGPK. |
| Molecular Weight : | 42.2 kDa (378aa) confirmed by MALDI-TOF. |
| Form : | Liquid. In 20 mM Tris-HCl buffer (pH8.0) containing 1mM DTT, 10% glycerol. |
| Purity : | > 90% by SDS – PAGE. |
| Concentration : | 1 mg/ml (determined by Bradford assay). |
| Storage : | Can be stored at +4°C short term (1-2 weeks). For long term storage, aliquot and store at -20°C or -70°C. Avoid repeated freezing and thawing cycles. |
| Gene Name | SAE1 SUMO1 activating enzyme subunit 1 [ Homo sapiens ] |
| Synonyms | SAE1; SUMO1 activating enzyme subunit 1; SUMO-1 activating enzyme E1 N subunit; SUMO-1 activating enzyme subunit 1; SUMO-activating enzyme subunit 1; activator of SUMO1; sentrin/SUMO-activating protein AOS1; ubiquitin-like 1-activating enzyme E1A; ubiquitin-like protein SUMO-1 activating enzyme; AOS1; FLJ3091; HSPC140 |
| Gene ID | 10055 |
| mRNA Refseq | NM_001145713 |
| Protein Refseq | NP_001139185 |
| MIM | 613294 |
| UniProt ID | Q9UBE0 |
| Chromosome Location | 19q13.32 |
| Pathway | Ubiquitin mediated proteolysis |
| Function | ATP-dependent protein binding; contributes_to SUMO activating enzyme activity; enzyme activator activity; ligase activity; protein heterodimerization activity; protein binding; ubiquitin activating enzyme activity |
| ◆ Recombinant Proteins | ||
| SAE1-292Z | Recombinant Zebrafish SAE1 | +Inquiry |
| SAE1-5223R | Recombinant Rat SAE1 Protein | +Inquiry |
| SAE1-1985H | Recombinant Human SAE1 protein | +Inquiry |
| SAE1-4889H | Recombinant Human SAE1 protein, GST-tagged | +Inquiry |
| SAE1-61H | Recombinant Human SUMO1 Activating Enzyme Subunit 1 | +Inquiry |
| ◆ Cell & Tissue Lysates | ||
| SAE1-001HCL | Recombinant Human SAE1 cell lysate | +Inquiry |
SUMOylation of E2F1 regulates expression of EZH2
Journal: Cancer research PubMed ID: 32816857 Data: 2022/1/7
Authors: Li Du, Marwan G. Fakih, Yuan Chen
Article Snippet:In CDH1 promoter luciferase assay, both WT and 4KR EZH2 showed similar dose-dependent suppression of the CDH1 promoter ( ).similar dose-dependent suppression of the CDH1 promoter ( ). ... Taken together, these data suggest that SUMOylation regulates EZH2 function mainly through its expression and not through direct SUMO modification of EZH2. fig ft0 fig mode=article f1 fig/graphic|fig/alternatives/graphic mode="anchored" m1 Open in a separate window Figure 6. caption a7 caption a8 EZH2 SUMOylation doesn’t affect its histone methyltranferase activity, protein stability or its suppression of CDH1 promoter. (A) A representative blot of in vitro SUMOylation of purified PRC2 (Creative-Biomart) by incubating with recombinant SUMO E1 (SAE1/SAE2), SUMO E2 enzyme (UBC9), and SUMO1 or SUMO2, and without or with RanBP2 (an E3) at 30oC for 3 h or 16 h. EZH2 was detected by western blot. (B) A representative blot of in vitro histone methytransferase assay with PRC2 after PRC2 in vitro SUMOylation.. SUMOylated PRC2 from in vitro SUMOylation was incubated with or without SENP1 for 30 min, and then histone H3 and S-adenosyl methionine were added for 60 min. Then, western blots were performed to detect H3K27me3 level. (C) Top, schematic diagram indicating the three detected and one predicted SUMOylation sites of EZH2—K20, K307, K419 and K421.SUMOylated PRC2 from in vitro SUMOylation was incubated with..
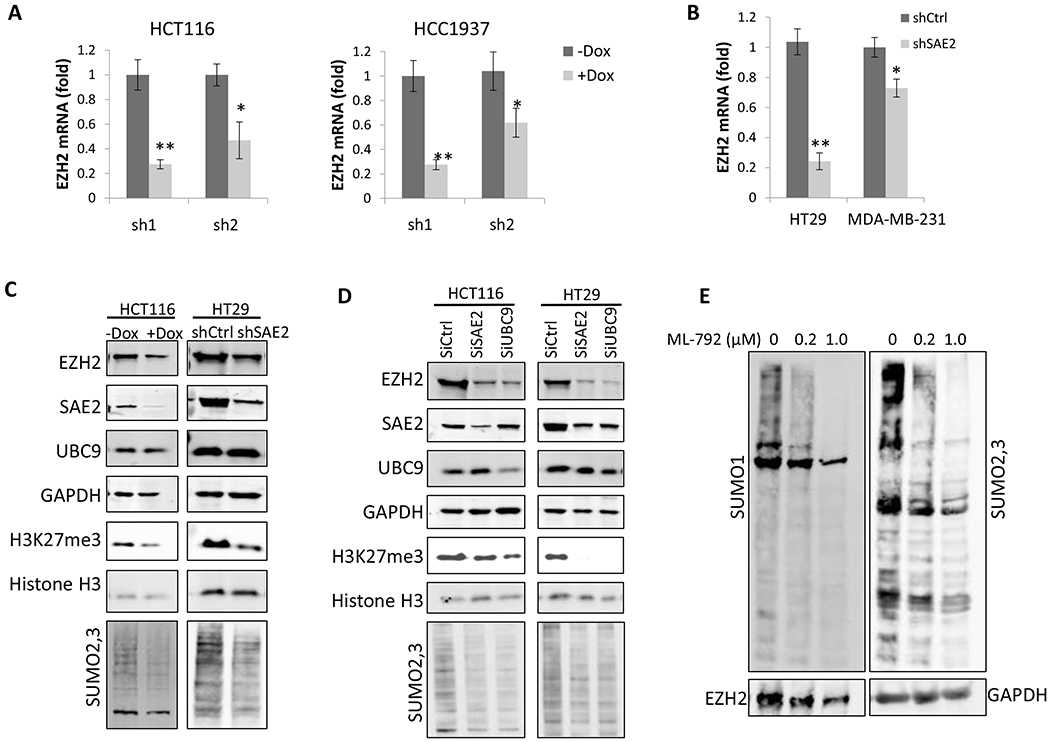
(A) EZH2 mRNA levels were determined using real-time qPCR in HCT116 cells (left panel) and HCC1937 cells (right panel). Doxycycline (Dox, 5μg/ml) was added to induce
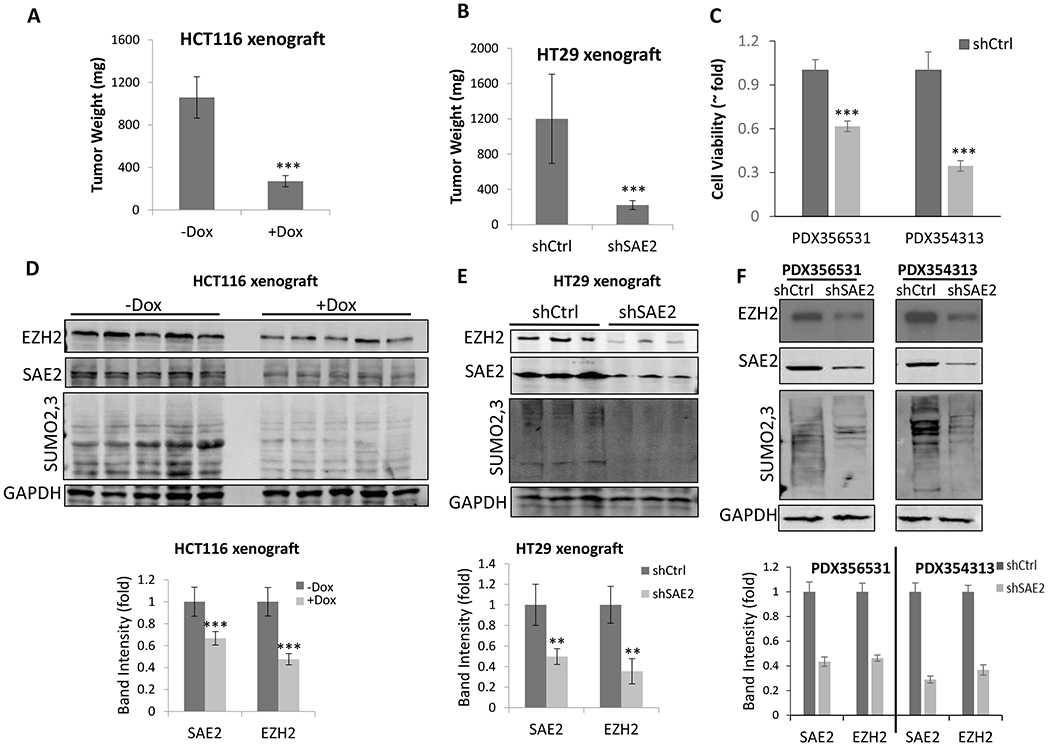
(A) Nude mice were xenografted with HCT116 cells transduced with Dox-inducible
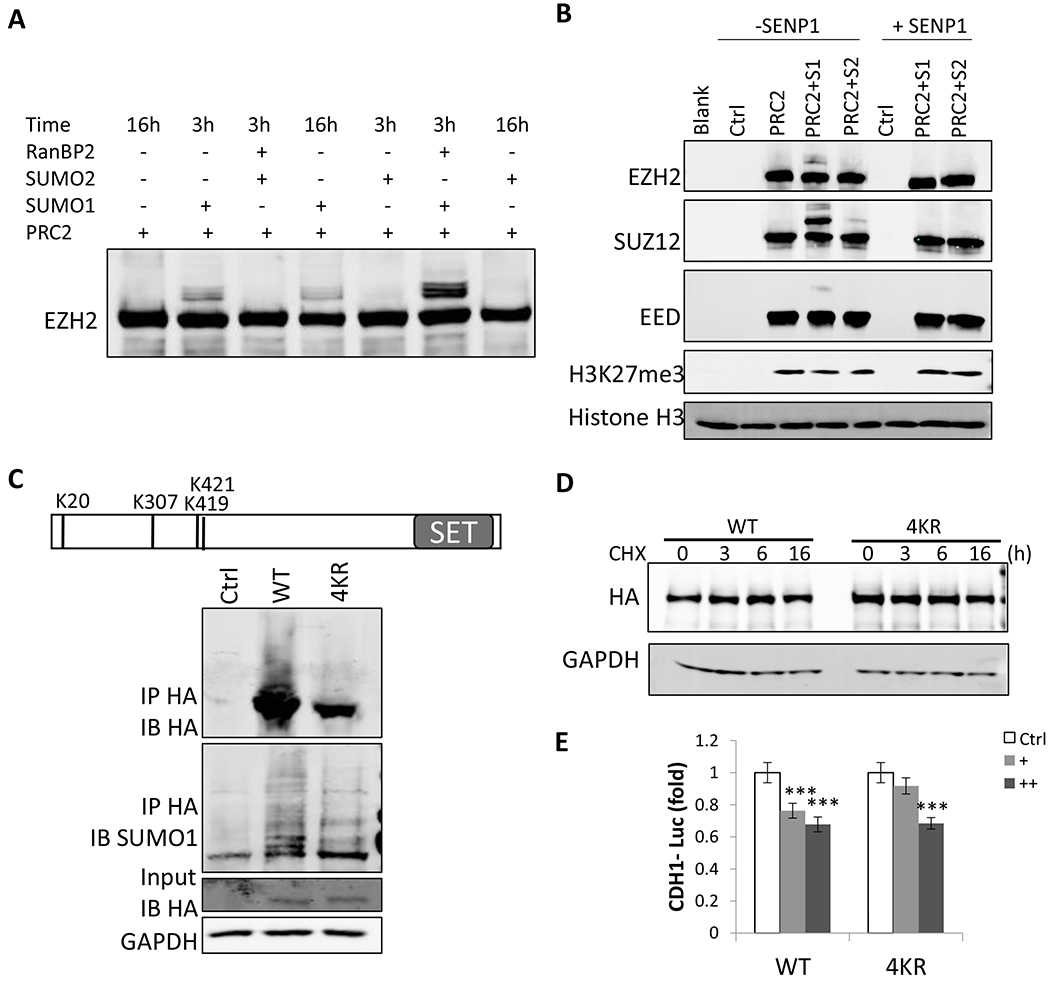
(A) A representative blot of in vitro SUMOylation of purified PRC2 (Creative-Biomart) by incubating with recombinant
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All SAE1 Products
Required fields are marked with *
My Review for All SAE1 Products
Required fields are marked with *



