Other Autophagy-related Proteins
Related Symbol Search List
- Bcl-2
- BCL2L1
- BNIP3
- BNIP3L
- C9orf72
- p27
- CIB1
- CELF1
- CHMP2B
- PFKFB3
- SIRT1
- SIRT2
- SMPD1
- SQSTM1
- TFEB
- TMEM59
- UVRAG
- VCP
Immunology Background
Autophagy is a cellular degradation system in which cytoplasmic components, including organelles, long-life proteins are sequestered by double-membrane structures called autophagosomes and the sequestered materials are degraded by lysosomal hydrolases. The process of autophagy is mainly divided into four parts, the induction activation of autophagy, the nucleation and extension of autophagosomes, the fusion of autophagosomes and lysosomes, and the degradation of contents.
The first step: the induction of autophagy.
A series of autophagy-related genes (Atg) is involved in the regulation of autophagosome formation, mainly the Atg1/ULK1 complex, including Atg1, Atg13, Atg17, mTORC1 phosphorylation of Atg1/ULK1 and Atg13 inhibit the initiation of autophagy.
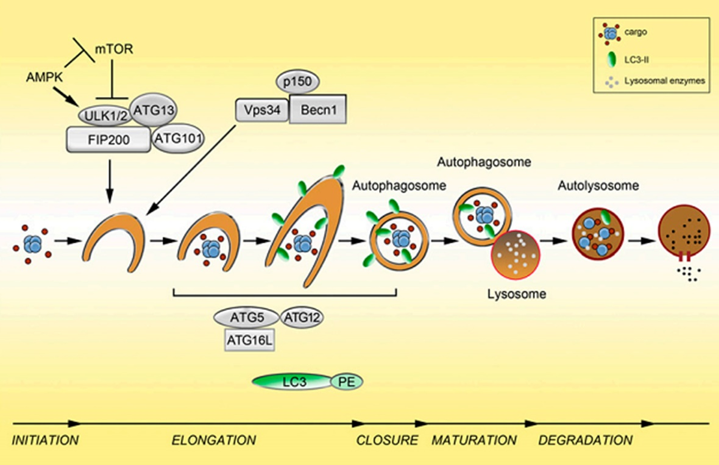
The second step: nucleation of autophagosomes.
After receiving the autophagy-inducing signal, the cells form a small membrane-like structure in the cytoplasm: the isolation membrane (IM), and then continuously extend to both sides, flattened, called pre-autophagosome (pre- Autophagosome PAS) is one of the signs of autophagy. Requires Atg6(Beclin1)-ClassIII PI3K complex and Atg12-Atg5-Atg16L complex.
Morphological characteristics: The autophagy precursor is mostly crescent-shaped or semi-annular, which is a free double-layer membrane structure with a cavity between the two membranes. As the autophagy precursor grows and wraps around the contents, the lumen becomes inconspicuous. A variety of ATG proteins are involved in the assembly of autophagy precursors. Component source: derived from a unique membrane structure formed independently or a membrane structure such as mitochondria, endoplasmic reticulum or Golgi complex.
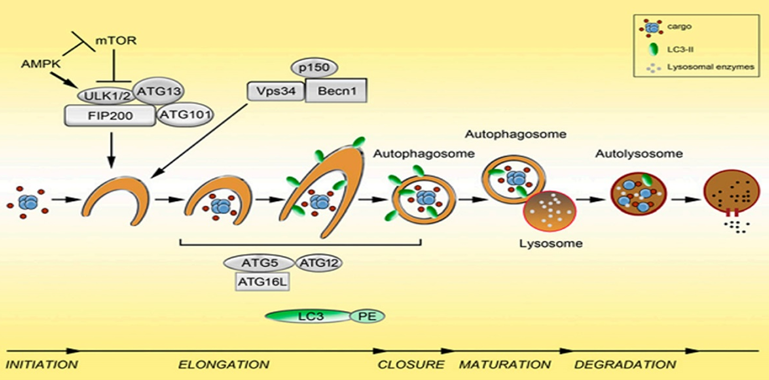
The extension of the autophagosome membrane, the autophagy precursor under the action of two ubiquitin-like systems, including the Atg12-Atg5-Atg16L complex and LC3 (Atg8), LC3 continuously accepts PE, expands the length of the membrane, and becomes mature. Close the autophagic bubble.
Morphological characteristics: The autophagy precursor consists of a half-moon shape, which continuously extends into a closed circular membrane of autophagic vacuoles. The contents are continuously captured proteins, organelles and protein aggregates to be degraded.
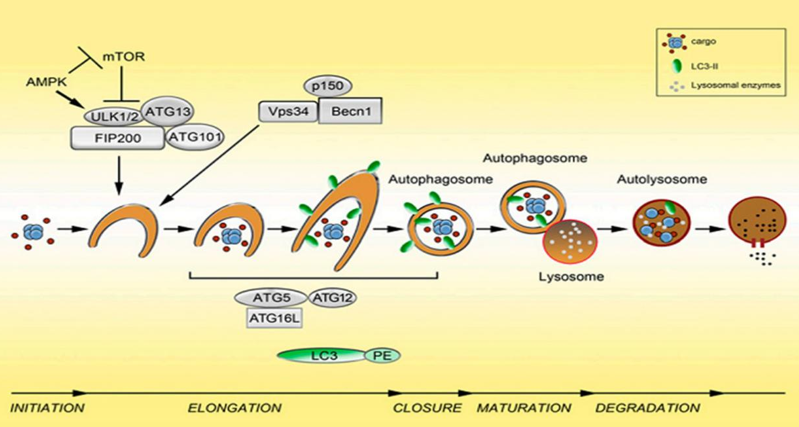
The third step: the fusion of autophagosomes and lysosomes is transported to lysosomes, and the biofilms of the two fuse to become an autophagic lysosome.
Morphological characteristics: mostly round or oval. Since the autophagy inner membrane is rapidly degraded, autophagic lysosomes are often not easily distinguished from other forms of lysosomes by transmission electron microscopy.
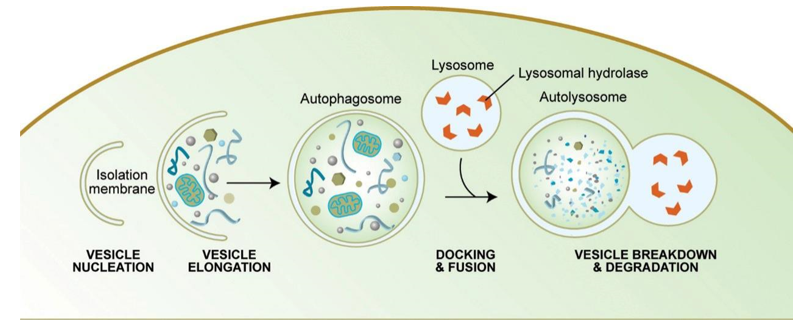
The fourth step: the acidic hydrolase in the autophagolysin lysate degrades the contents of the vesicle and becomes a degradative autophagic vacuole.
Regulation of autophagy
TOR (target of rapamycin), the central molecule of autophagy regulation
The target of rapamycin (TOR) is a receptor for amino acids, ATP and hormones, which plays an important role in regulating cell growth and autophagy. TOR (target of rapamycin) can sense a variety of changes in cells, enhance or reduce the level of autophagy. Intracellular ATP levels, hypoxia and other cellular signals can be directly or indirectly integrated by TOR, thereby changing the autophagy of cells and responding to different external environmental stimuli.
Atg1/ULK1 protein kinase complex
Atg1 is a serine/threonine protein kinase. The mammalian homologous protein ULK1, ULK1 exists as a complex. In addition to ULK1 itself, it also includes mAbg13, FIP200 (a protein that interacts with focal adhesion kinase FAK). And Atg101.When nutrients are sufficient, the mammalian rapamycin target protein complex 1 (mTORC1) binds to the Atg1/ULK1 complex and inhibits autophagy by phosphorylating Atg1/ULK1 and ATG13 thereby inhibiting the initiation of autophagy. Under starvation conditions, mTORC1 is isolated from the Atg1/ULK1 complex, thereby inducing nucleation and elongation of autophagosomes.
AMPK (AMP-activated protein kinase)
AMPK is a protein kinase that regulates the metabolism of energy states in cells and plays an important role in the regulation of autophagy. At low ATP levels (such as starvation or hypoxia), AMPK can activate the level of AMP and activate it, thereby phosphorylating the tuberous sclerosis protein (TSC2) to enhance the inhibition of Rheb by TSC1/2, ultimately making mTOR Activity is inhibited, and autophagy is induced in the cells.
Atg5-Atg12-Atg16 complex
There are two sets of ubiquitination modification processes in the process of autophagy, which occur in the Atg5-Atg12-Atg16 junction system and the Atg8/LC3 junction system, respectively, for the extension of the membrane and the formation of autophagic vacuoles. In the Atg5-Atg12-Atg16 junction system, Atg12 is first activated by Atg7 (E1); Atg12 is then transferred to the class Atg10 (E2). Finally, Atg12 is transferred to Atg5 and covalently bound to Lys of Atg5 to form Atg12-Atg5 complex. At the onset of autophagy, Atg16 binds to Atg12-Atg5, and Atg16 contains a helically coiled structure that tends to form oligomers that allow more of Atg12-Atg5 to join to form a giant complex. It is thus possible to provide a platform for interaction of proteins involved in autophagic bubble formation. When autophagy occurs, the complex is localized on the separator and participates in the formation of LC3-II, thereby promoting the elongation of the autophagic membrane.
Atg8/LC3
LC3 is a homolog of the yeast Atg8 molecule in mammals and is a marker protein for autophagy.
Function: Promote the elongation and fusion of autophagic membranes, which are necessary for the formation of autophagic vacuoles. After LC3 synthesis, it was initially in the form of pro-LC3 and was immediately cleaved by Atg4B to form LC3-I. When autophagy occurs, LC3-I uniformly distributed in the cytoplasm is activated by Atg7 (E1) and forms a thioester bond with it, which is then transferred to Atg3 (E2) and finally to Atg5-Atg12-Atg16 complex (E3). With the help of a previous PE molecule, LC3-II with membrane binding ability is formed. The cellular localization of Atg16L determines the location of LC3 maturation and promotes the binding of LC3-II to autophagic vacuoles.
ATG6/Beclin-1
The nucleation of autophagic vesicles requires a complex containing ATG6 (the mammalian homologous protein named Beclin 1), which forms a super complex with the third class of phosphoinositide 3 kinase VPS34 and activates it to produce phospholipids. Acyl inositol 3 phosphate. PI3P can assemble proteins containing PX and FYVE domains to the location of early autophagic vesicles, promoting the formation of autophagy precursors.
Vps34/PI3K-Beclin1 complex
Vps34 is a class III PI3K in mammals. In the Vps34 complex, Vps34 is activated by binding to Vps15 and further binds to Beclin1 to form the Vps34-Vps15-Beclin1 complex.
Function: When autophagy occurs, Vps34-Vps15-Beclin binds to a variety of autophagy-related proteins, and transmits autophagy signals to promote autophagy.
1. The Atg14-Vps34-Vps15-Beclin1 complex is formed by binding to Atg14 to participate in the formation of autophagic vacuoles.
2. Combined with VRAG to form VRAG-Vps34-Vps15-Beclin1 plays a role in autophagic vacuolar maturation and transport.
3. Promote the production of phosphatidylinositol-3-phosphate (PI3P), PI3P can assemble some proteins containing PX and FYVE domains to the location of early autophagic vesicles, and promote the formation of autophagy precursors.

