Recombinant Human TP53 Protein, Myc/DDK-tagged
| Cat.No. : | TP53-171H |
| Product Overview : | Recombinant Human TP53, transcript variant 1, fused with Myc/DDK tag at C-terminal was expressed in HEK293. |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Human |
| Source : | HEK293 |
| Tag : | DDK&Myc |
| Description : | This gene encodes a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. Mutations in this gene are associated with a variety of human cancers, including hereditary cancers such as Li-Fraumeni syndrome. Alternative splicing of this gene and the use of alternate promoters result in multiple transcript variants and isoforms. Additional isoforms have also been shown to result from the use of alternate translation initiation codons from identical transcript variants (PMIDs: 12032546, 20937277). |
| Form : | 25mM Tris-HCl, pH 7.3, 100mM glycine, and 10% glycerol. |
| Molecular Mass : | 43.6 kDa |
| Purity : | > 80% as determined by SDS-PAGE and Coomassie blue staining |
| Concentration : | >50 ug/mL as determined by microplate BCA method |
| Gene Name | TP53 tumor protein p53 [ Homo sapiens ] |
| Official Symbol | TP53 |
| Synonyms | TP53; tumor protein p53; cellular tumor antigen p53; LFS1; Li Fraumeni syndrome; p53; antigen NY-CO-13; mutant p53 protein; phosphoprotein p53; p53 tumor suppressor; truncated p53 protein; tumor suppressor TP53; transformation-related protein 53; P53; TRP53; FLJ92943; |
| Gene ID | 7157 |
| mRNA Refseq | NM_000546 |
| Protein Refseq | NP_000537 |
| MIM | 191170 |
| UniProt ID | P04637 |
| ◆ Recombinant Proteins | ||
| TP53-30547TH | Recombinant Human TP53, GST-tagged | +Inquiry |
| TP53-01HFL | Active Recombinant Full Length Human TP53 Protein, Flag tagged | +Inquiry |
| TP53-983H | Recombinant Human TP53 Protein, His-tagged | +Inquiry |
| TP53-5887R | Recombinant Rat TP53 Protein, His (Fc)-Avi-tagged | +Inquiry |
| TP53-6494H | Recombinant Human TP53 Protein (Full Length), N-His tagged | +Inquiry |
| ◆ Cell & Tissue Lysates | ||
| TP53-860HCL | Recombinant Human TP53 293 Cell Lysate | +Inquiry |
Development of cell-based high throughput luminescence assay for drug discovery in inhibiting OCT4/DNA-PKcs and OCT4–MK2 interactions
Journal: Biotechnology and bioengineering PubMed ID: 33565603 Data: 2021/5/1
Authors: Ismail S. Mohiuddin, Sung-Jen Wei, Min H. Kang
Article Snippet:About 100 units of purified DNA-PKcs (Promega) or 200 ng His-MK2 was incubated at 30°C for 30 min in 20 μl; kinase buffer containing 40 mM Tris (pH = 7.5), 20 mM MgCl 2 , 0.1 mg/ml BSA, activation buffer (100 μg/ml calf thymus DNA in 1× TE buffer), 150 μM ATP, and inhibitors of interest.100 units of purified DNA-PKcs (Promega) or 200 ng His-MK2 was incubated at 30°C for 30 min in 20 μl; kinase buffer containing 40 mM Tris (pH = 7.5), 20 mM MgCl 2 , 0.1 mg/ml BSA, activation buffer (100 μg/ml calf thymus DNA in 1× TE buffer), 150 μM ATP, and inhibitors of interest. ... Following the 30-min incubation, 1 μg of bacterially derived OCT4 (ProteinOne), p53 (Creative BioMart), and HSP27 (Enzo) protein substrates were added to the reaction.. Samples were incubated again for 30 min at 30°C, and then the reactions were quenched with the addition of 4× NuPAGE LDS and 100 mM dithiothreitol (DTT) before proceeding with immunoblotting, as described above.Samples were incubated again for 30 min at 30°C, and then the reactions were quenched with the addition of 4× NuPAGE LDS and 100 mM dithiothreitol (DTT) before proceeding with immunoblotting, as described above.
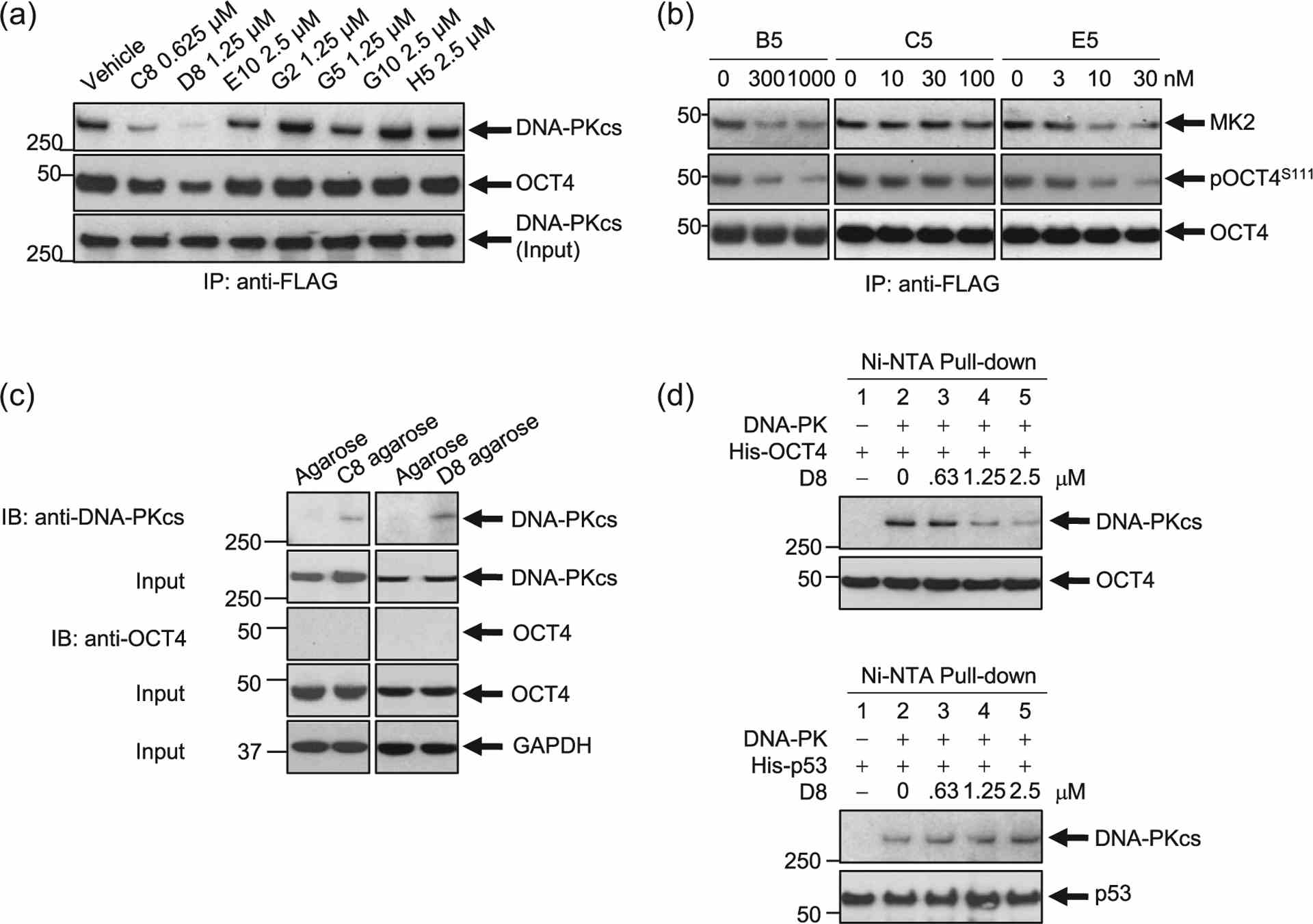
Hit Validation #2. Inhibition of the DNA-PKcs/OCT4 or MK2–OCT4 interaction following inhibitor treatment. (a) A stable clone of NCI-H82 cells expressing a DOX-inducible OCT4-mycDDK construct was treated with doxycycline for 18 h to induce OCT4-mycDDK expression. Cells were then treated with the inhibitors at the concentrations indicated for 6 h. Protein lysates were pulled down using EZView? Red Anti-FLAG? Affinity Gel. Immunoblotting was done to determine the effects of inhibitor treatment on the DNA-PKcs/OCT4 interaction. Two compounds, C8 and D8, decreased the expression of both DNA-PKcs and OCT4. Total DNA-PKcs expression input is assessed. (b) Hit validation by pull-down studies in the DOX-inducible OCT4-overexpressing SCLC cell line demonstrates that treatment with B5, C5, and E5 disrupt the protein–protein interaction between OCT4 and MK2 in a dose-dependent manner. (c) Stable clones of NCI-H82 cells expressing a DOX-inducible OCT4-mycDDK construct were treated with doxycycline for 18 h to induce OCT4-mycDDK expression. Protein lysates were pulled-down using C8- or D8-conjugated agarose. C8 and D8 bound to DNA-PKcs, not OCT4. (d) In vitro Ni-NTA pull-down assays. DNA-PKcs was incubated with D8 for 30 min before the addition of bacterially derived OCT4 and
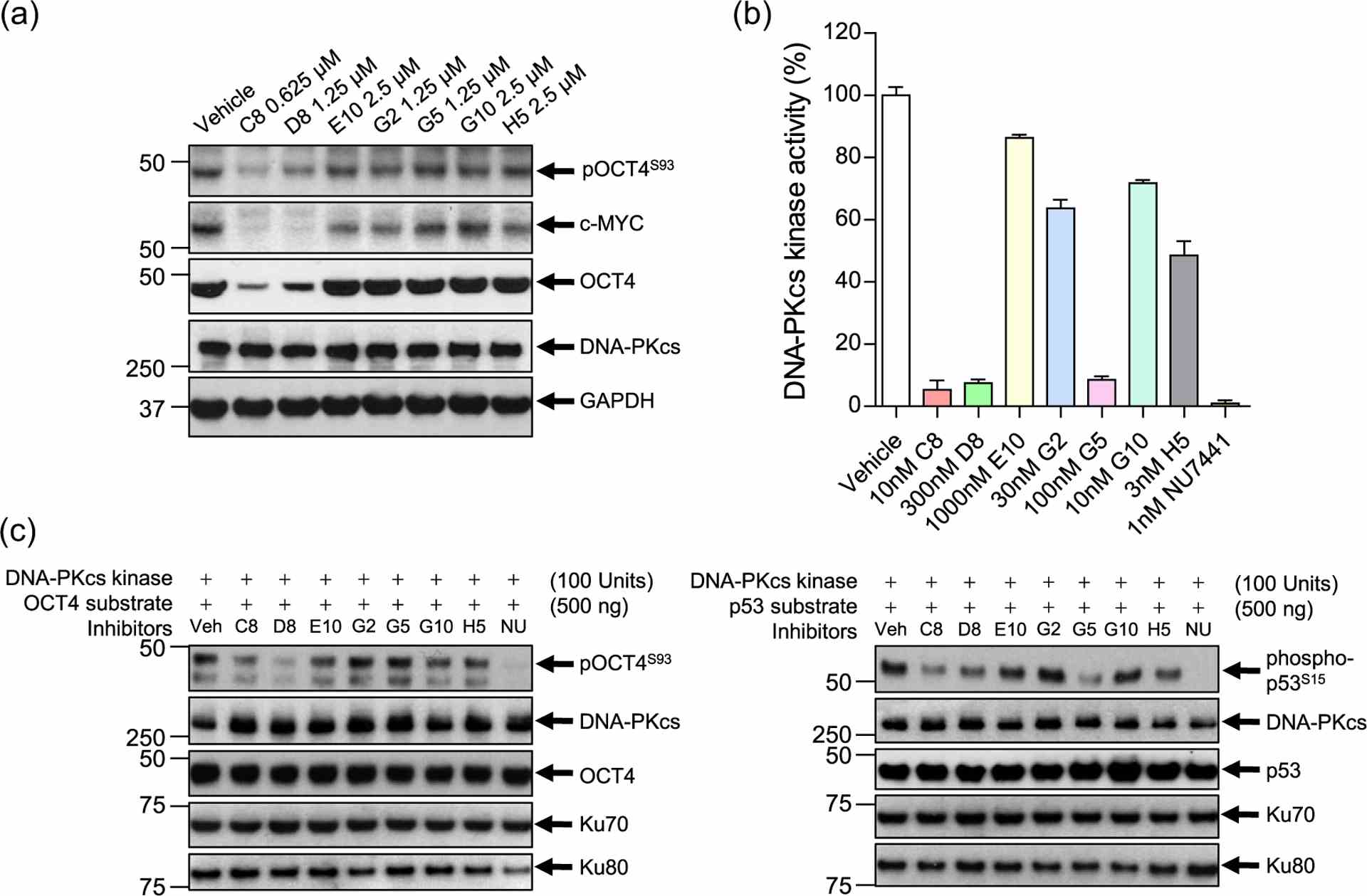
Hit validation, in vitro assays, and kinase activity assays for DNA-PKcs hits. (a) Hit validation demonstrating that two compounds: C8 and D8, inhibit OCT4, pOCT4S93, and c-MYC expression, but not DNA-PKcs, in a DOX-inducible OCT4-overexpressing SCLC cell line (NCI-H82 pCW57.1-OCT4-mycDDK). (b) ADP-Glo? assay demonstrating that C8, D8, and G5 drastically impair DNA-PKcs kinase activity. NU7441 (brown), a known DNA-PKcs inhibitor, was used as a positive control. (c) In vitro kinase activity assays. DNA-PKcs was incubated with inhibitors for 30 min before the addition of bacterially derived OCT4 (His-tag) and
Imaging and kinetics of the bimolecular complex formed by the tumor suppressor p53 with ubiquitin ligase COP1 as studied by atomic force microscopy and surface plasmon resonance
Journal: International Journal of Nanomedicine PubMed ID: 29379285 Data: 2022/12/14
Authors: Ilaria Moscetti, Anna Rita Bizzarri, Salvatore Cannistraro
Article Snippet:Tips were rinsed in DMSO to remove the unbound PEG.Tips were rinsed in DMSO to remove the unbound PEG.. Successively, they were incubated with 10 μL of p53 (5 μM, Creative BioMart) in PBS buffer overnight at 4°C, and they were gently rinsed with 10 mM PBS buffer and Milli-Q water.. 2D-Aldehyde-functionalized glass surfaces, 1 cm 2 (PolyAn GmbH, Berlin, Germany), characterized by a thin silane layer able to covalently bind proteins via their exposed amino groups, were incubated with 20 μL of COP1 (2.5 μM) in PBS buffer overnight at 4°C as described in .2D-Aldehyde-functionalized glass surfaces, 1 cm 2 (PolyAn GmbH, Berlin, Germany), characterized by a thin silane layer able to covalently bind proteins via their exposed amino groups, were incubated with 20 μL of COP1 (2.5 μM) in PBS buffer overnight at 4°C as described in .
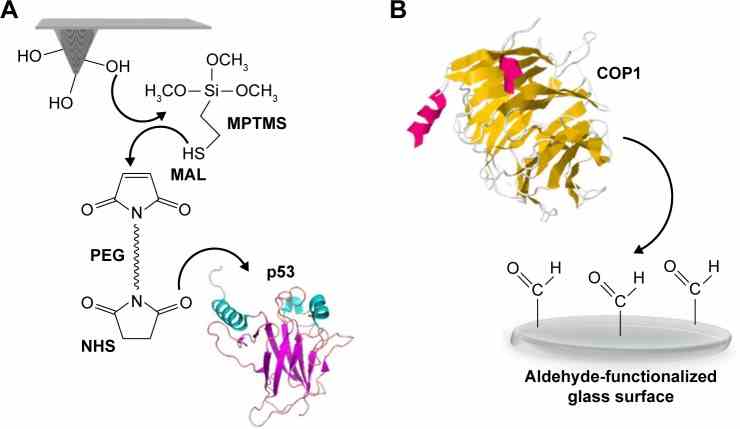
Schematic representation of the surface chemistry used to covalently bind
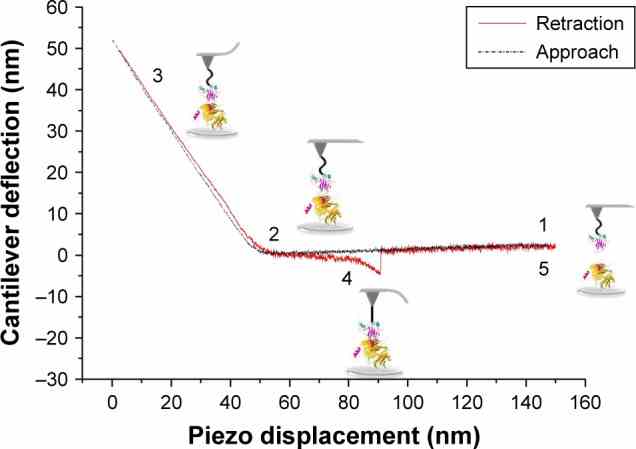
A typical approach–retraction cycle of the
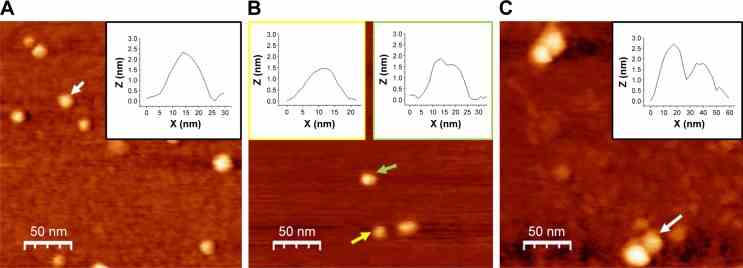
TM-AFM images recorded in air of COP1,
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All TP53 Products
Required fields are marked with *
My Review for All TP53 Products
Required fields are marked with *



