Hematopoietic Stem Cell Markers
Related Symbol Search List
- CD45
- SPI1
- CD244
- CD44
- CD93
- CD34
- ENG
- Kdr
- PODXL
- CD38
- KIT
- ABCG2
- ALDH1A1
- BMI1
- CD48
- CDCP1
- CXCR4
- ESAM
- GATA2
- ITGA6
- Mcl-1
- MECOM
- MYB
- PROCR
- PROM2
- PTEN
- Thy1
- TRAPPC4
- VCAM1
- ZBTB16
Immunology Background
Overview of Hematopoietic Stem Cell Markers
Hematopoietic stem cell markers refer to a group of cell surface molecules that are widely used as tools for identifying, isolating, and purifying hematopoietic stem cells (HSCs) in cell phenotyping and laboratory studies. Through an in-depth study of HSC markers, we can better understand the properties and functions of HSCs. Many markers have been identified for the identification and isolation of HSCs, such as CD34, CD38, CD45RA, CD133, Tie2, CD144, CD90 (Thy-1), CD117 (c-Kit), CD49f, EPCR/CD201, and RET. Among them, CD34 is a glycoprotein that is mainly expressed in the hematopoietic cell lines at early differentiation stages, such as hematopoietic stem cells and promyelocytes. CD133 is a glycoprotein cell surface molecule that is mainly expressed in neural tissues during the embryonic period, whereas it is predominantly expressed in hematopoietic and progenitor cells in adult tissues. CD38 is a cell surface ligand whose expression is significantly increased in mature hematopoietic cells. In summary, the study of hematopoietic stem cell markers enables us to better understand and utilize hematopoietic stem cells and provides important support for developing the field of stem cell therapy and regenerative medicine. Through further research and application, we expect to develop more accurate and efficient markers and promote the clinical application of hematopoietic stem cells.
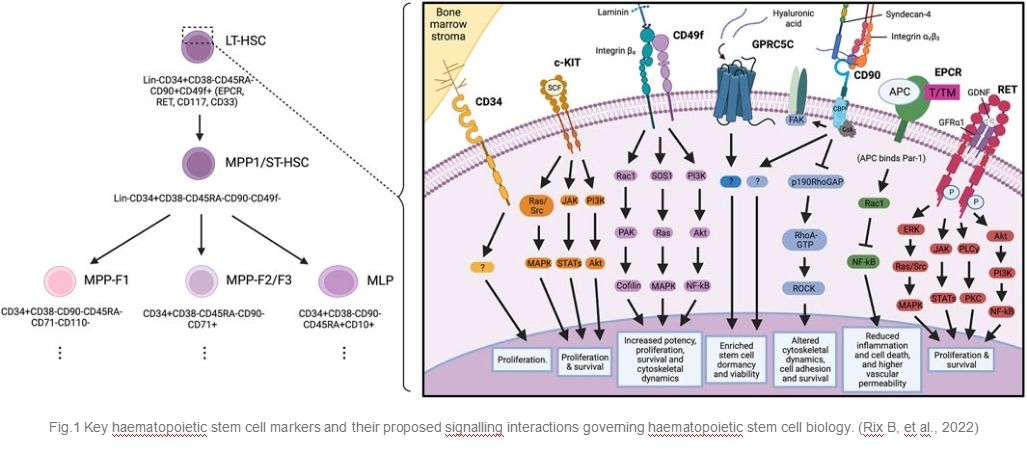 Fig.1 Key haematopoietic stem cell markers and their proposed signalling interactions governing haematopoietic stem cell biology. (Rix B, et al., 2022)
Fig.1 Key haematopoietic stem cell markers and their proposed signalling interactions governing haematopoietic stem cell biology. (Rix B, et al., 2022)Functions of Hematopoietic Stem Cell Markers
- Identification and separation of HSCs
The primary function of HSC markers is to aid in identifying and isolating HSCs with stem cell properties. HSCs can be differentiated and isolated from other cell types through the specific expression of surface markers. For example, CD34 and CD38 are often used for the initial identification of HSCs, while markers such as CD45, CD90, and CD133 are used to further subdivide and purify HSC subpopulations.
- To study the self-renewal and proliferation capacity of HSCs
By analyzing the expression patterns of markers and functional experiments, it is possible to understand which markers play a key role in the proliferation and stemness status of HSCs. For example, one study found that CD150 (Slamf1) and CD41 play an important role in the proliferation and self-renewal of HSCs.
- To study HSC differentiation and development
By observing the expression patterns and functions of markers, it is possible to reveal the differentiation mechanisms and regulatory factors of HSCs to different hematopoietic cell lines. For example, CD41 and CD45 play important roles in HSC differentiation and platelet formation (Miyawaki K, et al., 2015).
- Clinical applications
HSC markers can be used to diagnose and monitor blood-related diseases, evaluate the efficacy of HSCT, and develop new therapeutic strategies. For example, CD34 is widely used in hematopoietic stem cell transplantation for HSC selection and transplantation (Nielsen JS, et al., 2009).
- Research on the mechanisms of hematologic diseases
The prospect of studying blood hematopoietic stem cell markers is very promising. Through in-depth study and understanding of these markers and their relationship with hematopoietic stem cells, we are expected to further enhance the control and utilization of hematopoietic stem cells for better clinical applications and therapeutic effects. This will lay the foundation for potential treatments and possible cures for many diseases.
Through in-depth study and understanding of blood hematopoietic stem cell markers and their relationship to hematopoietic stem cells, we are expected to further enhance the control and utilization of hematopoietic stem cells for better clinical applications and therapeutic outcomes.
References
- Rix B, Maduro A H, Bridge K S, et al. Markers for human hematopoietic stem cells: The disconnect between an identification marker and its function[J]. Frontiers in physiology, 2022: 1906.
- Pang W W, Price E A, Sahoo D, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age[J]. Proceedings of the national academy of sciences, 2011, 108(50): 20012-20017.
- Liang Y, Van Zant G, Szilvassy SJ. Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood. 2005;106(4):1479-1487.
- Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Hematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(7186): 442-447.
- Miyawaki K, Arinobu Y, Iwasaki H, et al. CD41 marks the initial myelin-erythroid lineage specification in adult mouse hematopoiesis: redefinition of murine common myeloid progenitor. Stem cells. 2015;33(3):976-987.
- Nielsen JS, McNagny KM. CD34 is a key regulator of hematopoietic stem cell trafficking to bone marrow and mast cell progenitor trafficking in the periphery. Microcirculation. 2009;16(6):487-496.
- Felfly H, Haddad G G. Hematopoietic stem cells: potential new applications for translational medicine[J]. Journal of stem cells, 2014, 9(3): 163.

