Notch Signal Pathway
🧪 DYRK1A-19H
Source: Insect Cells
Species: Human
Tag: GST
Conjugation:
Protein Length:
🧪 AP2A2-9715H
Source: E.coli
Species: Human
Tag: His
Conjugation:
Protein Length: 590-939a.a.
🧪 BCL6-10185H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 183-532a.a.
🧪 CNTN1-11411H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 25-350a.a.
🧪 CSNK2B-11635H
Source: E.coli
Species: Human
Tag:
Conjugation:
Protein Length: 1-215 aa
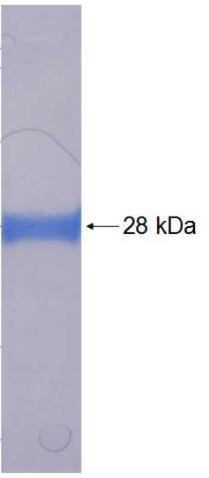
🧪 DLK1-12023H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 83-383a.a.
🧪 DNM1-12103H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 502-851a.a.
🧪 DNM2-12105H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 517-866a.a.
🧪 DNM3-12106H
Source: E.coli
Species: Human
Tag: His
Conjugation:
Protein Length: 206-555 aa
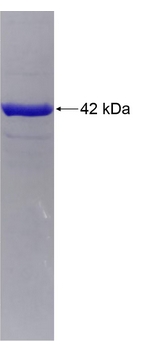
🧪 FURIN-13036H
Source: E.coli
Species: Human
Tag: His
Conjugation:
Protein Length: C-term-300a.a.
🧪 HDAC1-13703H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 183-482 aa

🧪 HDAC2-13706H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: C-term-299a.a.
🧪 IGLC1-14119H
Source: E.coli
Species: Human
Tag: His
Conjugation:
Protein Length: 233a.a.
🧪 MAML2-718H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: 805-1153aa
🧪 MIB1-870H
Source: E.coli
Species: Human
Tag: GST
Conjugation:
Protein Length: C-term-300aa
Background of Notch Signal Pathway
What is Notch Signal Pathway?
In 1917, Morgan and colleagues found a gene in Drosophila, which is named Notch because of a loss of function that leads to a gap in the edge of wings. Subsequent studies found that Notch is expressed in many species from invertebrates to vertebrates. The structure of Notch families is highly conserved and plays a key role in cell differentiation and development.
The Notch signaling pathway is composed of Notch receptors (Notch1-4), ligands (such as Delta-like 1, 3, 4, Jagged1 and Jagged2), transcription factor RBPJ (CBF-1, Suppressor of hairless, Lag) and downstream effector molecular composition. Activation of Notch signaling usually begins with the binding of Notch ligands to Notch receptors on the surface of neighboring cells. This binding triggers the Notch receptor to undergo two protease cleavages, eventually releasing Notch intracellular segments (NICD or ICN), which then migrate to the nucleus, bind to RBPJ and recruit coactivators such as MAML1 to form complexes that promote transcriptional activation of Notch target genes.
Up to now, studies have elucidated the major members and core transduction of Notch signaling pathway. However, as the research progresses, it is gradually realized that this pathway is actually in a very complicated regulatory network, which is consistent with its multiple function in development.
Core Components of Notch Signal Pathway
Notch Receptors
Notch receptor is a type I membrane protein with the relative molecular weight of about 30,000. There are four types of human Notch receptors: Notch1~4. It consists of extracellular and transmembrane subunits, and the two subunits form heterodimers via Ca2+ dependent non-covalent bonds. The extracellular subunit contains a set of tandemly arranged EGFR (epidermal growth factor receptor) and 3 family-specific LNR (Lin-12/Notch repeat) repeats. EGFR plays a pivotal role in the binding of Notch receptors to ligands. In Drosophila, the 11th and 12th EGFR of Notch receptors mediate their binding to ligands. LNR, located downstream of EGFR, is rich in cysteine and mediates a Ca2+ dependent interaction between the two subunits. Transmembrane subunits include transmembrane regions, RAM sequences, ankyrin (ANK) repeats, nuclear localization sequences, polyglutamine sequences, and PEST sequences (a peptide sequence that is rich in proline (P), glutamic acid (E), serine (S), and threonine (T)). The RAM domain is the major binding site for the Notch signaling effector CBF1 / RBPJκ. The ANK repeat domain is the binding site of Deltex and Mastermind. These proteins have a function of modification to the Notch signaling pathway. The PEST domain is associated with ubiquitin-mediated degradation of the Notch intracellular domain.

Figure 1. Structures of human Notch receptors.
Notch Ligands
Notch ligands are also type I transmembrane proteins. Drosophila Notch ligands have two homologues Delta and Serrate, the nematode Notch ligand is LAG2. Hence the Notch ligand is also known as DSL protein. A number of Notch ligands are also found in vertebrate, the Delta homology is called Delta-like molecules, and the Serrate homology is called Jagged. Currently, human Notch ligands are found to have DLL 1, 3, 4 and Jagged 1, 2. Ligand Extracellular DSL domains are highly conserved in evolution and are essential for ligand-receptor binding to activate Notch signaling. Intracellular domain of Notch ligands is short, only contains 70 amino acid residues, the function has not yet been clarified. Recent studies have found that the intracellular domain of Delta 1 can induce cell growth inhibition. It is speculated that the intracellular segment of the ligand may be similar with the receptor intracellular domain to have the function of signal transduction, but the specific mechanism needs further study.

Figure 2. Structures of human Notch ligands.
Notch Signaling and Effectors
The existing research proposed the "three-step proteolytic model" of Notch signaling activation. First, Notch is synthesized in the endoplasmic reticulum in a single-stranded precursor pattern, and then is cleaved in the Golgi apparatus by furin-like invertase into large fragments with a molecular mass of 180,000 extracellular regions, and a molecular mass of 120,000 transmembrane domains and small fragments of the intracellular region. These two fragments are combined to become heterodimers via Ca2+ dependent non-covalent bonds and then transported to the cell membrane. When Notch ligand binds to receptor, Notch receptor undergoes proteolysis twice. For the first time, Notch receptors were cut into two fragments by the ADAM metalloprotease family ADAM10 / Kuz or ADAM17 / TACE. The N-terminal cleavage product (extracellular region) is endocytosed by the ligand-expressing cells and the C-terminal cleavage product (active Notch receptor NICD) is subsequently enzymatically released by the γ-secretase complex consisting of presenilin 1/2, Pen-2, Aphl and Nicastrin.

Figure 3. Canonical Notch signal pathway.
The canonical Notch signal pathway is also known as CBF1 / RBPJκ dependent pathway. CBF1 / RBPJκ is a transcriptional repressor that binds specifically to the DNA sequence "CGTGGGAA" and recruits proteins such as SMRT, SKIP, and type I / II histone deacetylases to form costimulatory complexes and inhibit the transcription of downstream genes. After Notch signaling is activated, NICD is released into the nucleus by the enzyme cleavage reaction, and the co-suppression complex is dissociated by binding of the RAM domain and the ANK repeat to CBF1 / RBPJK and recruitment of SKIP and MAML1 to form the co-active complex and activate the transcription of downstream genes. Many of the target genes for Notch signaling are members of the basic helix-loop-helix transcriptional repressor family, such as HES in mammals, XHey-1 in Xenopus laevis, and recently discovered BLBPs. In addition, a CBF1 / RBPJK independent Notch signal transduction exists. Recently, it has been reported that Drosophila Notch-binding protein Deltex is required for certain tissue-specific non-Su (H) dependent signals and that Deltex also has antagonistic function against Notch.
Notch Signal Pathway Related Diseases
Notch signaling pathway has a very conservative mechanism in signal transduction, the abnormality of this regulatory mechanism often leads to congenital genetic diseases. It has been confirmed that the mutations of related genes in the Notch signaling pathway are associated with genetic diseases such as CADASIL, Aligile's syndrome and hypogastric hypoplasia. CADASIL is an autosomal dominant cerebral artery disease with subdural necrosis and leukoencephalopathy. Aligile's syndrome is an autosomal dominant genetic disease that can lead to the developmental defects of many organs such as heart, liver and kidney. In addition, the study found that Notch signaling disorders associated with certain cardiovascular diseases. Animal model experiments show that it may affect the cardiovascular system from four aspects, including vascular remodeling, vascular stability, choice of arteriovenous and heart development. Recent studies have found that the cleavage of amyloid precursor protein and Notch receptor are all dependent on γ-secretase / presenilin. So it is speculated that Notch signaling pathway may have some connection with the occurrence and development of Alzheimer's disease, and AD related research confirmed the change of Notch signal.

Figure 4. Notch signaling in disease. (Chris Siebel, 2017)
It is also expected that the signal transduction abnormality will lead to the tumor. This relationship was first confirmed in human acute T-lymphoblastic leukemia. Subsequent studies found that in prostate cancer, breast cancer, cervical cancer and other tumor cells, as well as their derived cell lines, there are abnormal expression of Notch receptors and ligands. In addition to mutations in Notch receptors and ligands that can cause tumors, mutations in cofactor MAML that mediate association of NICD with CSL in the signaling pathway can also lead to tumorigenesis. In addition, Notch signaling can interact with other signaling pathways to induce tumors. However, it was found that Notch also plays a role of tumor suppressor, which also reflects the cell microenvironment closely related to the specific function of Notch signaling.
Case Study
Case Study 1: Recombinant Human JAG1 (JAG1-3138H)
Bone marrow-derived mesenchymal stem cell-based treatments, which do not have the side effects of BMP2, are not currently FDA approved, and are time and resource intensive. There is a critical need for novel bone regenerative therapies to treat CF bone loss that have minimal side effects, are easily available, and are affordable. In this study we investigated novel bone regenerative therapies downstream of JAGGED1 (JAG1). We previously demonstrated that JAG1 induces murine cranial neural crest (CNC) cells towards osteoblast commitment via a NOTCH non-canonical pathway involving JAK2-STAT5 (1) and that JAG1 delivery with CNC cells elicits bone regeneration in vivo. In this study, the researchers hypothesized that delivery of JAG1 and induction of its downstream NOTCH non-canonical signaling in pediatric human osteoblasts constitute an effective bone regenerative treatment in an in vivo murine bone loss model of a critically-sized cranial defect. Using this CF defect model in vivo, we delivered JAG1 with pediatric human bone-derived osteoblast-like (HBO) cells to demonstrate the osteo-inductive properties of JAG1 in human cells and in vitro we utilized the HBO cells to identify the downstream non-canonical JAG1 signaling intermediates as effective bone regenerative treatments. In vitro, we identified an important mechanism by which JAG1 induces pediatric osteoblast commitment and bone formation involving the phosphorylation of p70 S6K. This discovery enables potential new treatment avenues involving the delivery of tethered JAG1 and the downstream activators of p70 S6K as powerful bone regenerative therapies in pediatric CF bone loss.

Fig1. Alizarin Red S dye was extracted from stained cells using a 1:10 dilution of acetic acid and water, and the absorbance was read at 420nm. (5.7 μM). (Archana Kamalakar, 2023)
Case Study 2: Recombinant Human DLL1 (DLL1-218H)
An emerging dogma shows that tumors are initiated and maintained by a subpopulation of cancer cells that hijack some stem cell features and thus referred to as "cancer stem cells" (CSCs). The exact mechanism that regulates the maintenance of CSC pool remains largely unknown. Fascin is an actin-bundling protein. Here, the researchers manipulated fascin expression in breast cancer cell lines and used several in vitro and in vivo approaches to examine the relationship between fascin expression and breast CSCs. Fascin knockdown significantly reduced stem cell-like phenotype (CD44hi /CD24lo and ALDH+ ) and reversal of epithelial to mesenchymal transition. Interestingly, expression of the embryonic stem cell transcriptional factors (Oct4, Nanog, Sox2, and Klf4) was significantly reduced when fascin expression was down-regulated. Functionally, fascin-knockdown cells were less competent in forming colonies and tumorspheres, consistent with lower basal self-renewal activity and higher susceptibility to chemotherapy. Fascin effect on CSC chemoresistance and self-renewability was associated with Notch signaling. Activation of Notch induced the relevant downstream targets predominantly in the fascin-positive cells.

Fig2. Bar graph showing relative RNA expression. Cells were treated with Notch ligand (DLL1). (Rayanah Barnawi, 2016)
Case Study 3: Recombinant Human NOTCH1 protein
Notch signaling, a critical pathway for tissue development, contributes to tumorigenesis in many tissues; however, the roles of Notch signaling in Intrahepatic Cholangiocarcinoma (ICC) remains unclear. This study evaluated the expression and effects of Notch1 on cell migration in ICC. Multiple cellular and molecular approaches were performed including gene transfection, siRNA transfection, RT-PCR, Western blotting, Rac activation assays and immunofluorescence. The results showed that the exogenous expression of Notch1 in glioma cells increased their migratory and invasive capacity. Similarly, the suppression of Notch1 expression inactivated Rac1 and inhibited ICC cell migration. Notch1 over expression induced an Epithelial-to-mesenchymal transition (EMT) phenotype that included enhanced expression of α-SMA and Vimentin, loss of E-cadherin expression, morphological changes and cytoskeletal reorganization in ICC cells.

Fig3. Notch1 overexpression induces an EMT phenotype in ICC-9810 cells. (Qi Zhou, 2013)
Related Products
The Notch signaling pathway is an evolutionarily highly conserved cellular communication mechanism, found in both non-vertebrates and vertebrates, and it plays an important role in cell growth, fate determination, embryogenesis, and cell proliferation and differentiation. The main task of future research is to unveil the mechanisms of the complex regulatory networks of Notch signaling, thereby recognizing the basis for the diversity of functions of Notch signaling that will allow us to design more specific approaches and treatment program to specific pathologies resulting from aberrant Notch signaling. Creative BioMart can provide related protein products of the Notch signal pathway to help your researches. Please feel free to contact us if you’re interested.


