Native Calf Histone
| Cat.No. : | Histone-52C |
| Product Overview : | Mixture of histones H1, H2A, H2B, H3, and H4, isolated from calf thymus. |
- Specification
- Gene Information
- Related Products
- Case Study
- Application
- Download
| Species : | Calf |
| Source : | Calf thymus |
| Description : | istones are a group of DNA-binding proteins that are characterized by relatively high levels of lysine and arginine. Five different fractions of histones have been isolated and characterized. These are named as H1, H2A, H2B, H3 and H4. The H1 fraction is lysine-rich, while the H2A and H2B are slightly lysine-rich, The H3 and H4 fractions are arginine-rich. The molecular weights of histones are approximately 11 to 21 kDa depending on the fraction. |
| Form : | Lyophilizate |
| Bio-activity : | Histones have an important role in the organization and modification of chromatin. The nucleosome is the basic unit of the chromatin. It is made up of two molecules each of histones H2A, H2B, H3 and H4, wrapped with approximately 146 bp of DNA. Histone H1 binds to the nucleosomes at the DNA entry and exit points and stabilizes them. |
| Applications : | Histone from calf thymus has been used: in in vitro kinase assay to check the phosphorylation of histones by protein kinase A. to check the in vitro methyltransferase activity of protein argininemethyltransferases from Oryza sativa (OsPRMTs). in anti-histone ELISA. |
| Notes : | For life science research only. Not for use in diagnostic procedures. |
| Stability : | Aqueous solution is stable at -15 to -25 centigrade for several months. |
| Storage : | 2-8 centigrade. |
| Shipping : | Wet ice |
| Reconstitution : | Recommended solvent is double-distilled water. |
| Official Symbol | Histone |
| Synonyms | Histone |
| ◆ Native Proteins | ||
| Histone-52C | Native Calf Histone | +Inquiry |
| Histone-53C | Native Calf Histone Protein | +Inquiry |
Case 1: Kysela B, et al. Proc Natl Acad Sci U S A. 2005
The DNA-dependent protein kinase (DNA-PK) and DNA ligase IV/XRCC4 (LX) complexes are necessary for DNA nonhomologous end-joining in vivo. This study investigated how histone octamers and linker histone H1 affect DNA end-joining in vitro. Here forming dinucleosomes from the DNA substrate doesn't significantly hinder LX ligation, but the presence of linker histones greatly reduces it. This inhibition is independent of core histone octamers, cannot be overcome by Ku alone, but is partially reversed by DNA-PK, which efficiently phosphorylates histone H1. Phosphorylated H1 has less affinity for DNA, reducing its inhibitory effect on end-joining. This suggests DNA-PK might function as a linker histone kinase, phosphorylating linker histones near DNA breaks to facilitate localized histone H1 release and enable LX recruitment for ligation. Using histone H1-bound DNA, we successfully reconstituted the end-joining step in vitro, demonstrating a requirement for DNA-PK.
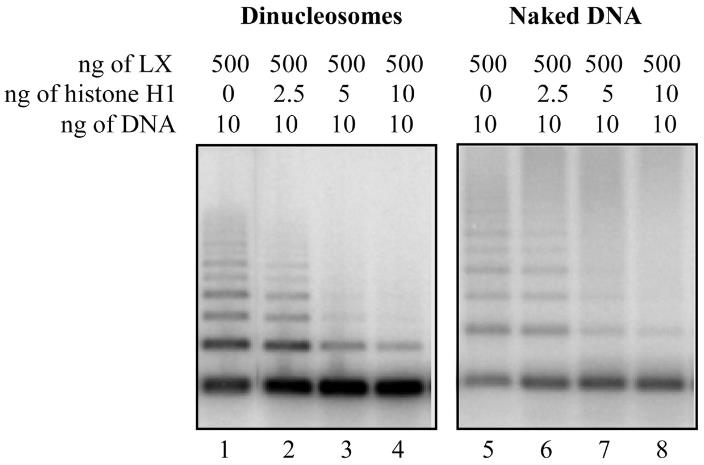
Fig1. Histone H1 inhibits LX ligation in the presence or absence of histone octamers.
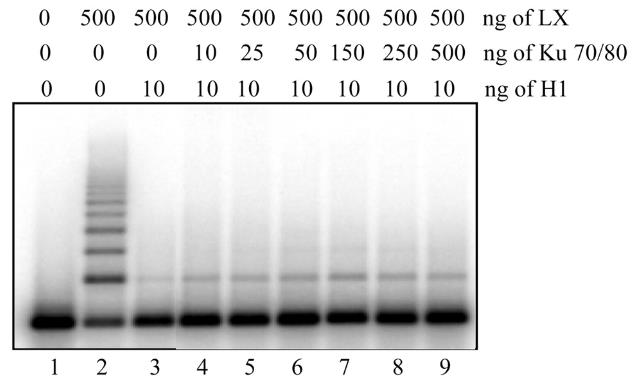
Fig2. Addition of Ku does not overcome the histone H1-dependent inhibition of LX ligation.
Case 2: Saumer P, et al. Nucleic Acids Res. 2024
Post-translational modifications (PTMs) of histones, including linker histone H1, significantly influence chromatin structure and function. However, the understanding of H1 modifications is limited by the absence of suitable tools. In this study, researchers created chromatosomes with site-specifically modified H1.2 using chemical biology methods. These were then analyzed using affinity enrichment mass spectrometry to identify interactomes and the effects of H1 modifications at a proteome level. They confirmed the findings by western-blotting and showed that chromatin-bound H1.2 aids in recruiting DNA repair proteins using an in vitro assay. The data expands on previous studies by providing insights into modification-specific interactions of H1, particularly when bound to chromatin.
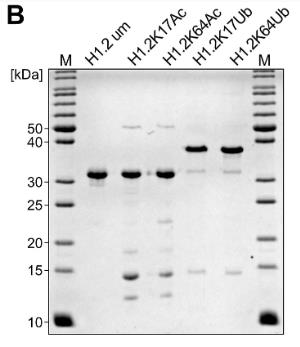
Fig1. Schematic overview and SDS-PAGE of all H1.2 variants generated for this study.
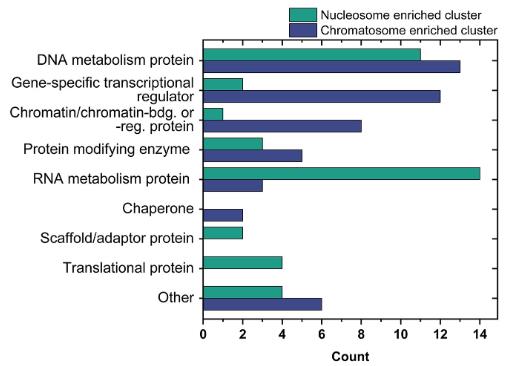
Fig2. Classification of enriched proteins by GO-terms using PANTHER for the nucleosome (green) and chromatosome (blue) cluster.
Native Calf Histone, extracted from calf thymus, consists of the primary nucleic acid-binding proteins found in the cell nuclei of eukaryotic organisms. These proteins play a pivotal role in the structure and function of chromatin by wrapping around DNA to form nucleosomes, which help compact DNA and regulate gene expression. The main types of histones include H1, H2A, H2B, H3, and H4, each with distinct roles in the chromatin complex.
In research, Native Calf Histone is extensively used for studying gene expression regulation, as histones are central to understanding how genes are turned on and off. It is also crucial for investigating the structure of chromatin and its dynamic role in the cell cycle, as well as its function in the transmission of genetic information. Furthermore, histones are targeted in drug development, with enzymes that modify histones, such as histone demethylases and acetyltransferases, being potential therapeutic targets, especially in cancer treatment. Additionally, since histone modifications are associated with various diseases like cancer and neurodegenerative disorders, Native Calf Histone is employed in disease research to explore pathogenesis and potential therapeutic strategies. Overall, Native Calf Histone is a valuable tool in biomedical research, and understanding its role and function can further advance scientific and clinical applications in related fields.
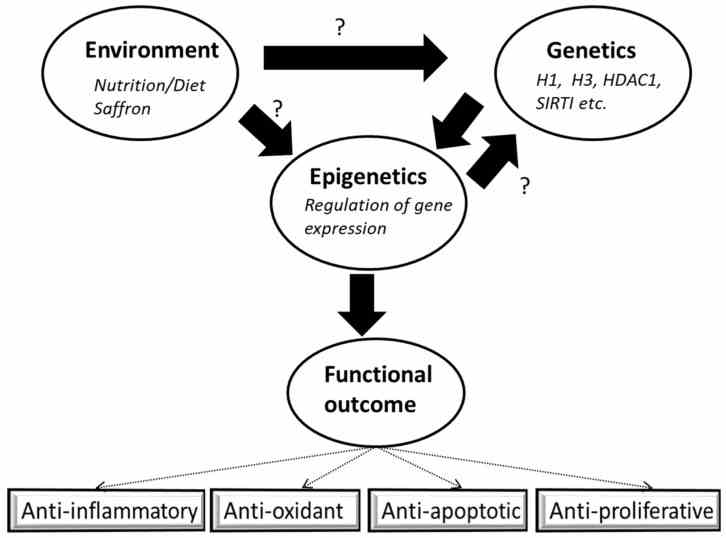
Fig1. Interplay of environmental factors with genetics and epigenetics. (Mudasir Rashid, 2022)
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All Histone Products
Required fields are marked with *
My Review for All Histone Products
Required fields are marked with *



