Active Recombinant Human IL10RA, HIgG1 Fc-tagged
| Cat.No. : | IL10RA-212H |
| Product Overview : | The extracellular domain of human IL-10RA (AAH28082.1) (His22-Asn235) is fused to the N-terminus of the Fc region of a human IgG1 was expressed in CHO cell. |
| Availability | February 01, 2026 |
| Unit | |
| Price | |
| Qty |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Human |
| Source : | CHO |
| Tag : | Fc |
| Protein Length : | 22-235 a.a. |
| Description : | The protein encoded by this gene is a receptor for interleukin 10. This protein is structurally related to interferon receptors. It has been shown to mediate the immunosuppressive signal of interleukin 10, and thus inhibits the synthesis of proinflammatory cytokines. This receptor is reported to promote survival of progenitor myeloid cells through the insulin receptor substrate-2/PI 3-kinase/AKT pathway. Activation of this receptor leads to tyrosine phosphorylation of JAK1 and TYK2 kinases. Two transcript variants, one protein-coding and the other not protein-coding, have been found for this gene. |
| Form : | Lyophilized from 0.2μm-filtered solution in PBS. |
| Bio-activity : | Measured by its ability to inhibit IL-10-dependent proliferation of MC/9 2 mouse mast cells. |
| Molecular Mass : | 51KDa (monomer) |
| AA Sequence : | His22-Asn235 |
| Endotoxin : | <0.06 eu/μg="" as="" determined="" by="" lal="" test.=""> |
| Purity : | >98%, by SDS-PAGE under reducing conditions. |
| Stability : | Stable for at least 1 year after receipt when stored at -20°C. Working aliquots are stable for up to 3 months when stored at -20°C. |
| Reconstitution : | Reconstitute at 100μg/ml in sterile PBS. |
| Warning : | Avoid freeze/thaw cycles. |
| Publications : |
|
| Gene Name | IL10RA interleukin 10 receptor, alpha [ Homo sapiens ] |
| Official Symbol | IL10RA |
| Synonyms | IL10RA; interleukin 10 receptor, alpha; IL10R; interleukin-10 receptor subunit alpha; CD210; CD210a; CDW210A; HIL 10R; IL-10RA; IL-10R subunit 1; IL-10R subunit alpha; IL-10 receptor subunit alpha; interleukin-10 receptor subunit 1; interleukin-10 receptor alpha chain; HIL-10R; IL-10R1; |
| Gene ID | 3587 |
| mRNA Refseq | NM_001558 |
| Protein Refseq | NP_001549 |
| MIM | 146933 |
| UniProt ID | Q13651 |
| Chromosome Location | 11q23 |
| Pathway | Cytokine-cytokine receptor interaction, organism-specific biosystem; Cytokine-cytokine receptor interaction, conserved biosystem; Jak-STAT signaling pathway, organism-specific biosystem; Jak-STAT signaling pathway, conserved biosystem; Toxoplasmosis, organism-specific biosystem; Toxoplasmosis, conserved biosystem; Tuberculosis, organism-specific biosystem; |
| Function | interleukin-10 receptor activity; protein binding; receptor activity; signal transducer activity; |
| ◆ Recombinant Proteins | ||
| IL10RA-4323H | Recombinant Human IL10RA protein, His&Myc-tagged | +Inquiry |
| IL10RA-0297H | Active Recombinant Human IL10RA protein, Fc-Avi-tagged, Biotinylated | +Inquiry |
| IL10RA-6744H | Recombinant Human IL10RA protein, His-Avi-tagged | +Inquiry |
| IL10RA-9302H | Recombinant Human IL10RA protein, MYC/DDK-tagged | +Inquiry |
| IL10RA-2859H | Recombinant Human IL10RA protein(291-370 aa), C-His-tagged | +Inquiry |
| ◆ Cell & Tissue Lysates | ||
| IL10RA-1185CCL | Recombinant Cynomolgus IL10RA cell lysate | +Inquiry |
Investigation the Possibility of Using Peptides with a Helical Repeating Pattern of Hydro-Phobic and Hydrophilic Residues to Inhibit IL-10
Journal: PLoS ONE PubMed ID: 27100390 Data: 2016/4/21
Authors: Guoying Ni, Shu Chen, Massimiliano Galdiero
Article Snippet:Lipopolysaccharide (LPS) and Incomplete Freund’s adjuvant (IFA) were purchased from Sigma.Lipopolysaccharide (LPS) and Incomplete Freund’s adjuvant (IFA) were purchased from Sigma.. Recombinant Human interleukin 10 receptor alpha was purchased from Creative BioMart, USA (Cat. No IL10RA-212H), and was re-suspended in sterilized Milli Q water to a concentration of 1 μg/μL as stock solution.. Recombinant Human interleukin 5 receptor alpha was purchased from Genscript, USA (Cat. No Z03126-10), and was re-suspended in sterilized Milli Q water to a concentration of 1 μg/μL as stock solution.Recombinant Human interleukin 5 receptor alpha was purchased from Genscript, USA (Cat. No Z03126-10), and was re-suspended in sterilized Milli Q water to a concentration of 1 μg/μL as stock solution.
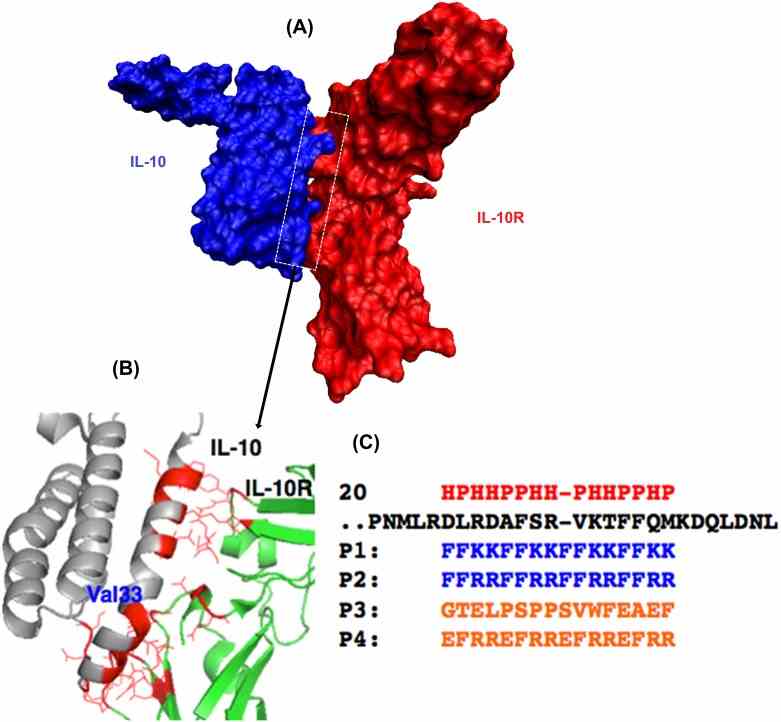
(A) A space-filling model of the interface structure of the IL-10/IL-10R protein–protein complex. (B) An expansion of the interface between the two proteins to indicate a few residue to residue contacts. The complex structure of IL-10 (Grey)
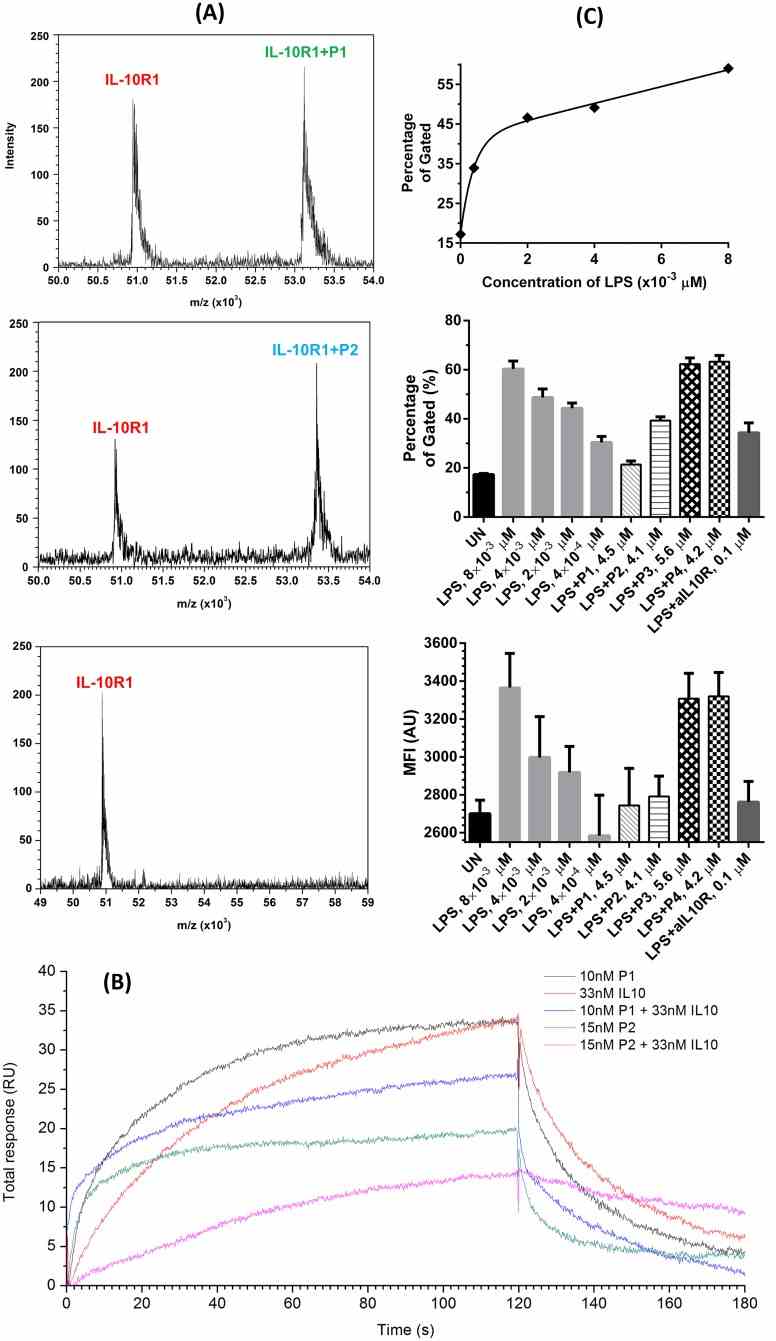
(A) MALDI mass spectra
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All IL10RA Products
Required fields are marked with *
My Review for All IL10RA Products
Required fields are marked with *



