Recombinant Human CDH1 protein, Fc-tagged
| Cat.No. : | CDH1-275H |
| Product Overview : | Recombinant Human CDH1 protein (Gln23-Pro621), fused to human IgG1 Fc tag at C-terminus, was expressed in human 293 cells (HEK293). |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Human |
| Source : | HEK293 |
| Tag : | Fc |
| Protein Length : | 599 |
| Description : | Cadherins are calcium-dependent cell adhesion proteins. They preferentially interact with themselves in a homophilic manner in connecting cells; cadherins may thus contribute to the sorting of heterogeneous cell types. Cadherin-1 (CDH1) is also known as epithelial cadherin (E-cadherin), CD_antigen (CD324), Uvomorulin (UVO) ECAD and CDHE, CDH1/CD324 contains 5 cadherin domains. CDH1 /CD324/ECAD is expressed in non-neural epithelial tissues. CDH1/E-CAD is involved in mechanisms regulating cell-cell adhesions, mobility and proliferation of epithelial cells and has a potent invasive suppressor role. It is a ligand for integrin alpha-E/beta-7. E-Cad promotes non-amyloidogenic degradation of Abeta precursors and has a strong inhibitory effect on APP C99 and C83 production. Defects in CDH1/CD324/ECAD are the cause of hereditary diffuse gastric cancer (HDGC). |
| Form : | Lyophilized from 0.22 um filtered solution in 50 mM Tris, 100 mM Glycine, pH7.5, 10% trehalose. |
| Molecular Mass : | The protein has a calculated MW of 92.4 kDa. The protein migrates as 100 kDa and 120 kDa Cadherin, a 48 kDa Propeptide under reducing (R) condition (SDS-PAGE) due to different glycosylation. |
| Endotoxin : | Less than 1.0 EU per ug by the LAL method. |
| Purity : | >95% as determined by SDS-PAGE. |
| Storage : | For long term storage, the product should be stored at lyophilized state at -20 centigrade or lower. Please avoid repeated freeze-thaw cycles. This product is stable after storage at: -20 centigrade to -70 centigrade for 12 months in lyophilized state; -70 centigrade for 3 months under sterile conditions after reconstitution. |
| Reconstitution : | It is recommended that sterile water be added to the vial to prepare a stock solution of 0.2 ug/ul. Centrifuge the vial at 4℃ before opening to recover the entire contents. |
| Gene Name | CDH1 |
| Official Symbol | CDH1 |
| Synonyms | CDH1; cadherin 1, type 1, E-cadherin (epithelial); UVO; cadherin-1; CD324; E Cadherin; uvomorulin; CAM 120/80; E-Cadherin; cell-CAM 120/80; epithelial cadherin; cadherin 1, E-cadherin (epithelial); calcium-dependent adhesion protein, epithelial; CDHE; ECAD; LCAM; Arc-1 |
| Gene ID | 999 |
| mRNA Refseq | NM_004360 |
| Protein Refseq | NP_004351 |
| MIM | 192090 |
| UniProt ID | P12830 |
| ◆ Recombinant Proteins | ||
| Cdh1-10529M | Recombinant Mouse Cdh1 Protein, His (Fc)-Avi-tagged | +Inquiry |
| CDH1-548H | Active Recombinant Human CDH1 Protein, Fc Chimera | +Inquiry |
| CDH1-214H | Recombinant Human CDH1 Protein, His-tagged | +Inquiry |
| CDH1-26935TH | Recombinant Human CDH1 | +Inquiry |
| CDH1-558H | Recombinant Human CDH1 Protein, His (Fc)-Avi-tagged | +Inquiry |
| ◆ Cell & Tissue Lysates | ||
| CDH1-1892MCL | Recombinant Mouse CDH1 cell lysate | +Inquiry |
| CDH1-938HCL | Recombinant Human CDH1 cell lysate | +Inquiry |
| CDH1-736RCL | Recombinant Rat CDH1 cell lysate | +Inquiry |
SUMOylation of E2F1 regulates expression of EZH2
Journal: Cancer research PubMed ID: 32816857 Data: 2022/1/7
Authors: Li Du, Marwan G. Fakih, Yuan Chen
Article Snippet:In CDH1 promoter luciferase assay, both WT and 4KR EZH2 showed similar dose-dependent suppression of the CDH1 promoter ( ).similar dose-dependent suppression of the CDH1 promoter ( ). ... Taken together, these data suggest that SUMOylation regulates EZH2 function mainly through its expression and not through direct SUMO modification of EZH2. fig ft0 fig mode=article f1 fig/graphic|fig/alternatives/graphic mode="anchored" m1 Open in a separate window Figure 6. caption a7 caption a8 EZH2 SUMOylation doesn’t affect its histone methyltranferase activity, protein stability or its suppression of CDH1 promoter. (A) A representative blot of in vitro SUMOylation of purified PRC2 (Creative-Biomart) by incubating with recombinant SUMO E1 (SAE1/SAE2), SUMO E2 enzyme (UBC9), and SUMO1 or SUMO2, and without or with RanBP2 (an E3) at 30oC for 3 h or 16 h. EZH2 was detected by western blot. (B) A representative blot of in vitro histone methytransferase assay with PRC2 after PRC2 in vitro SUMOylation.. SUMOylated PRC2 from in vitro SUMOylation was incubated with or without SENP1 for 30 min, and then histone H3 and S-adenosyl methionine were added for 60 min. Then, western blots were performed to detect H3K27me3 level. (C) Top, schematic diagram indicating the three detected and one predicted SUMOylation sites of EZH2—K20, K307, K419 and K421.SUMOylated PRC2 from in vitro SUMOylation was incubated with..
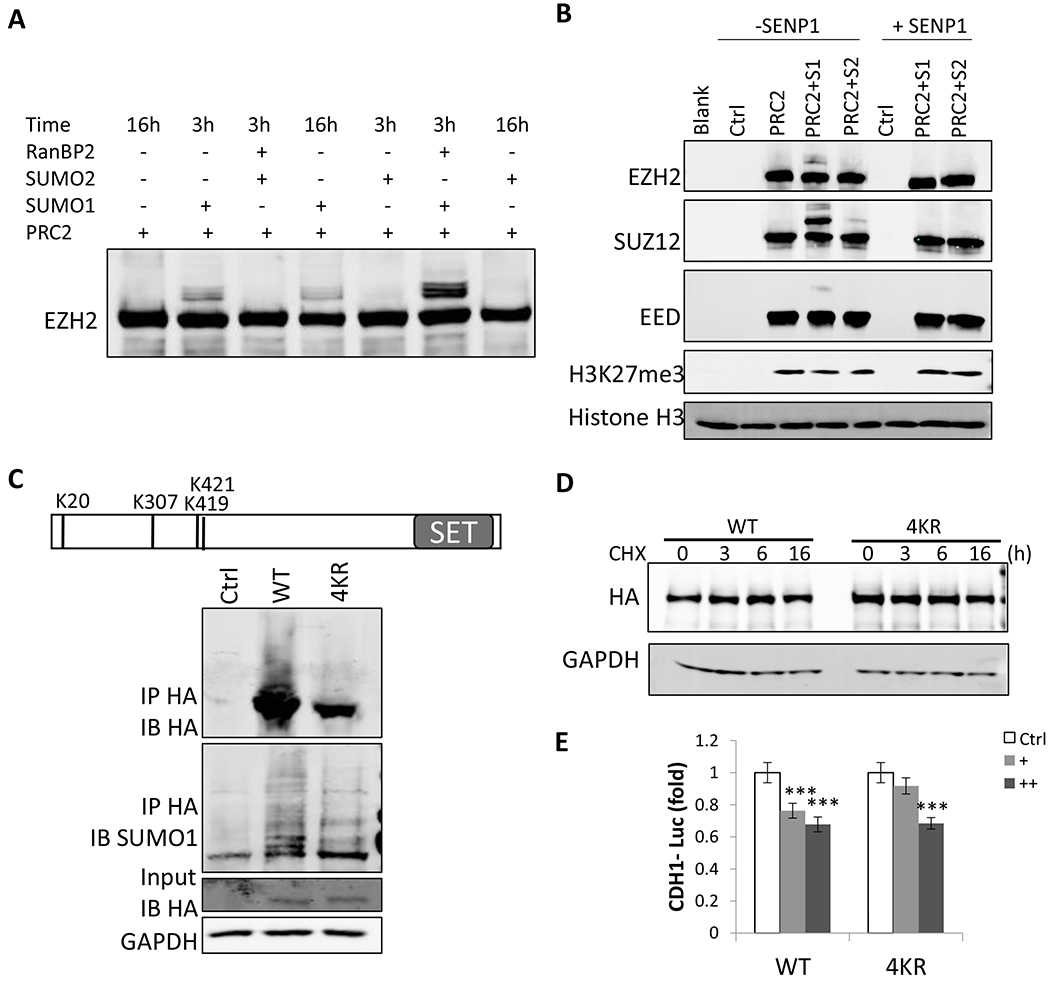
(A) A representative blot of in vitro SUMOylation of purified PRC2 (Creative-Biomart) by incubating with recombinant SUMO E1 (SAE1/SAE2), SUMO E2 enzyme (UBC9), and SUMO1 or SUMO2, and without or with RanBP2 (an E3) at 30oC for 3 h or 16 h. EZH2 was detected by western blot. (B) A representative blot of in vitro histone methytransferase assay with PRC2 after PRC2 in vitro SUMOylation. SUMOylated PRC2 from in vitro SUMOylation was incubated with or without SENP1 for 30 min, and then histone H3 and S-adenosyl methionine were added for 60 min. Then, western blots were performed to detect H3K27me3 level. (C) Top, schematic diagram indicating the three detected and one predicted SUMOylation sites of EZH2—K20, K307, K419 and K421. 293T cells were transfected with plasmids expressing HA-tagged WT or mutant EZH2 with 4 SUMOylation sites mutated to arginine (4KR) along with an UBC9-expressing vector. Then, the expression of both WT and mutant EZH2 were detected by western blots with an anti-HA antibody. Cell lysates were immunoprecipitated with an anti-HA antibody under denaturing condition followed by immunoblotting with an anti-HA antibody and SUMO1 antibody. (D) SUMOylation did not affect EZH2 protein stability. Representative western blot of EZH2 in HCT116 cells transfected with HA-tagged EZH2 WT or 4KR mutant expression plasmid for 2 days, followed by treatment with 100 μg ml?1 CHX for indicated time. GAPDH was used as a loading control. (E) SUMOylation doesn’t affect EZH2 function in suppressing
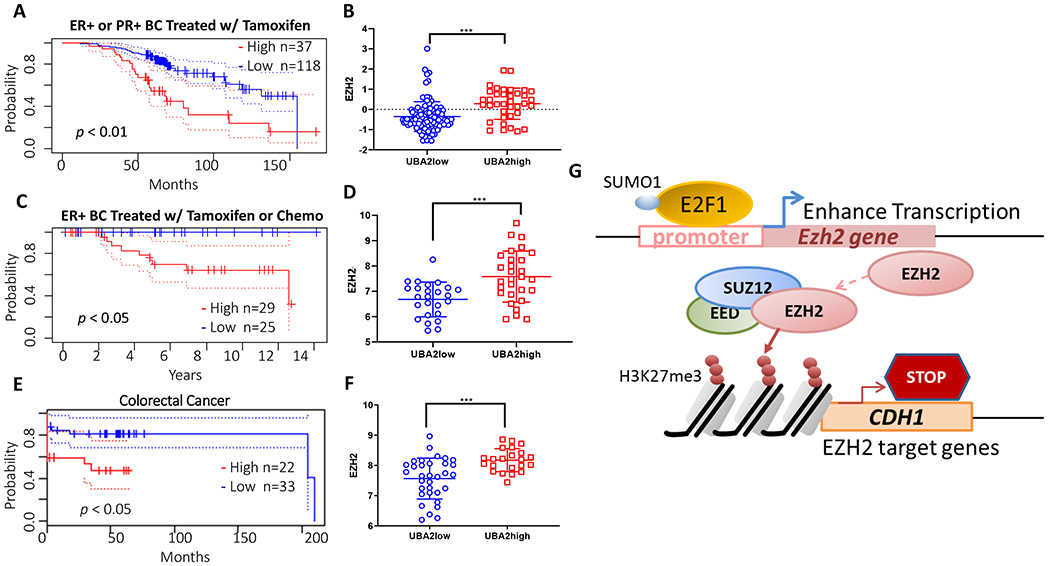
(A, B) Analysis for cohorts of 132 ER+ and/or PR+ breast cancer patients (GSE9893) treated with adjuvant tamoxifen therapy. (C, D) Analysis of cohort of 54 node-negative ER+ breast cancer patients (GSE7378) treated with tamoxifen or chemotherapy. The expression of SAE2 (UBA2) correlates to poor survival for both cohorts (A and C). Patients with high SAE2 (UBA2) (UBA2high group) showed higher EZH2 levels than patients with low SAE2 (UBA2) (UBA2low group) in both cohorts (B and D). A colon cancer cohort (GSE17537) also showed similar correlation (E and F). Kaplan-Meier plots were generated using online server Prognoscan. Solid lines represent two patient groups— high SAE2 level group (red) and low SAE2 level group (blue). Dotted lines indicate the 95% confidence intervals for each group. P values were derived using a two-tailed Student’s t-test. * p < 0.05, *** p < 0.001. (G) Schematic showing the mechanism of SUMOylation of E2F1 leads to increased EZH2 transcription, resulting in
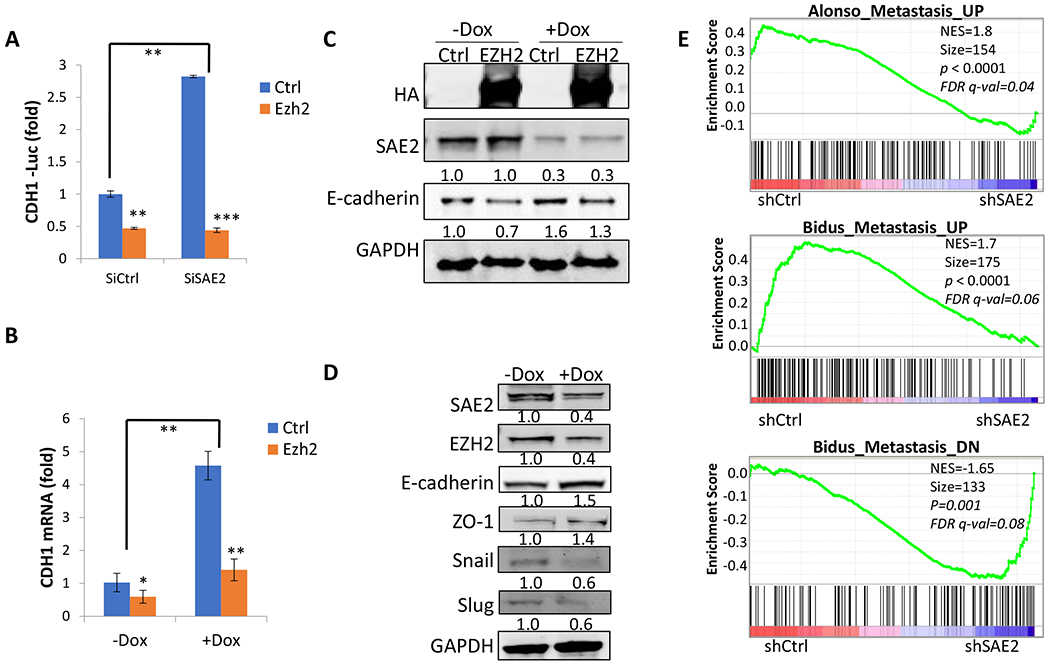
(A) Knockdown of SAE2 increased
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All CDH1 Products
Required fields are marked with *
My Review for All CDH1 Products
Required fields are marked with *



