Recombinant Rat SPI1 Protein
| Cat.No. : | SPI1-5706R |
| Product Overview : | Recombinant Rat SPI1 full length or partial length protein was expressed. |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Rat |
| Source : | Mammalian Cells |
| Tag : | His |
| Form : | Liquid or lyophilized powder |
| Endotoxin : | < 1.0 EU per μg of the protein as determined by the LAL method. |
| Purity : | >80% |
| Notes : | This item requires custom production and lead time is between 5-9 weeks. We can custom produce according to your specifications. |
| Storage : | Store it at +4 ºC for short term. For long term storage, store it at -20 ºC~-80 ºC. |
| Storage Buffer : | PBS buffer |
| Gene Name | Spi1 spleen focus forming virus (SFFV) proviral integration oncogene spi1 [ Rattus norvegicus ] |
| Official Symbol | SPI1 |
| Gene ID | 366126 |
| mRNA Refseq | NM_001005892.2 |
| Protein Refseq | NP_001005892.1 |
| MIM | |
| UniProt ID | Q6BDS1 |
| ◆ Recombinant Proteins | ||
| SPI1-172H | Recombinant Human SPI1 protein, His-tagged | +Inquiry |
| Spi1-4533M | Recombinant Mouse Spi1 protein, His&Myc-tagged | +Inquiry |
| SPI1-6580C | Recombinant Chicken SPI1 | +Inquiry |
| SPI1-5365R | Recombinant Rat SPI1 Protein, His (Fc)-Avi-tagged | +Inquiry |
| SPI1-5706R | Recombinant Rat SPI1 Protein | +Inquiry |
| ◆ Cell & Tissue Lysates | ||
| SPI1-1683HCL | Recombinant Human SPI1 cell lysate | +Inquiry |
Open Chromatin Profiling in Adipose Tissue Marks Genomic Regions with Functional Roles in Cardiometabolic Traits
Journal: G3: Genes|Genomes|Genetics PubMed ID: 31186305 Data: 2019/6/11
Authors: Maren E. Cannon, Kevin W. Currin, Karen L. Mohlke
Article Snippet:For EMSA, we prepared nuclear cell extracts from SGBS preadipocyte and SW872 cells using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific) as previously described. ( Kulzer et al. 2014 ) Double-stranded oligos (Table S12) were incubated with SGBS preadipocyte or SW872 nuclear extract or 100 ng purified PU.1 protein (Creative BioMart SPI1-172H) and DNA-protein complex visualization was carried out as previously described. ( Kulzer et al. 2014 ) A positive control oligo contained the PU.1 motif from JASPAR and a negative control did not contain the motif (Table S12).. We repeated all EMSA experiments on independent days and obtained consistent results.We repeated all EMSA experiments on independent days and obtained consistent results.
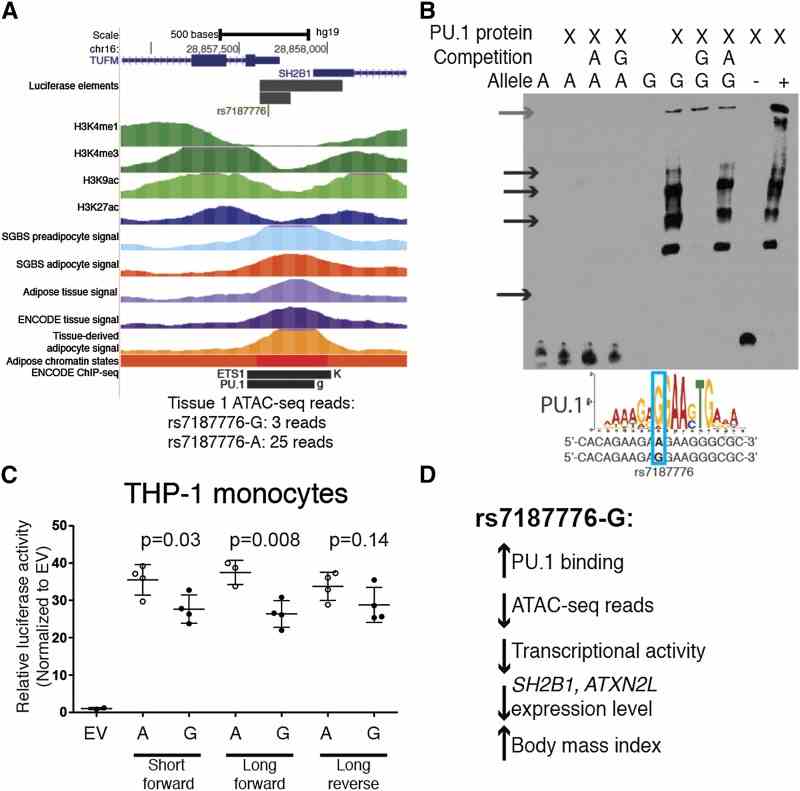
A variant at the ATP2A1-SH2B1 BMI GWAS locus alters chromatin accessibility and
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All SPI1 Products
Required fields are marked with *
My Review for All SPI1 Products
Required fields are marked with *



